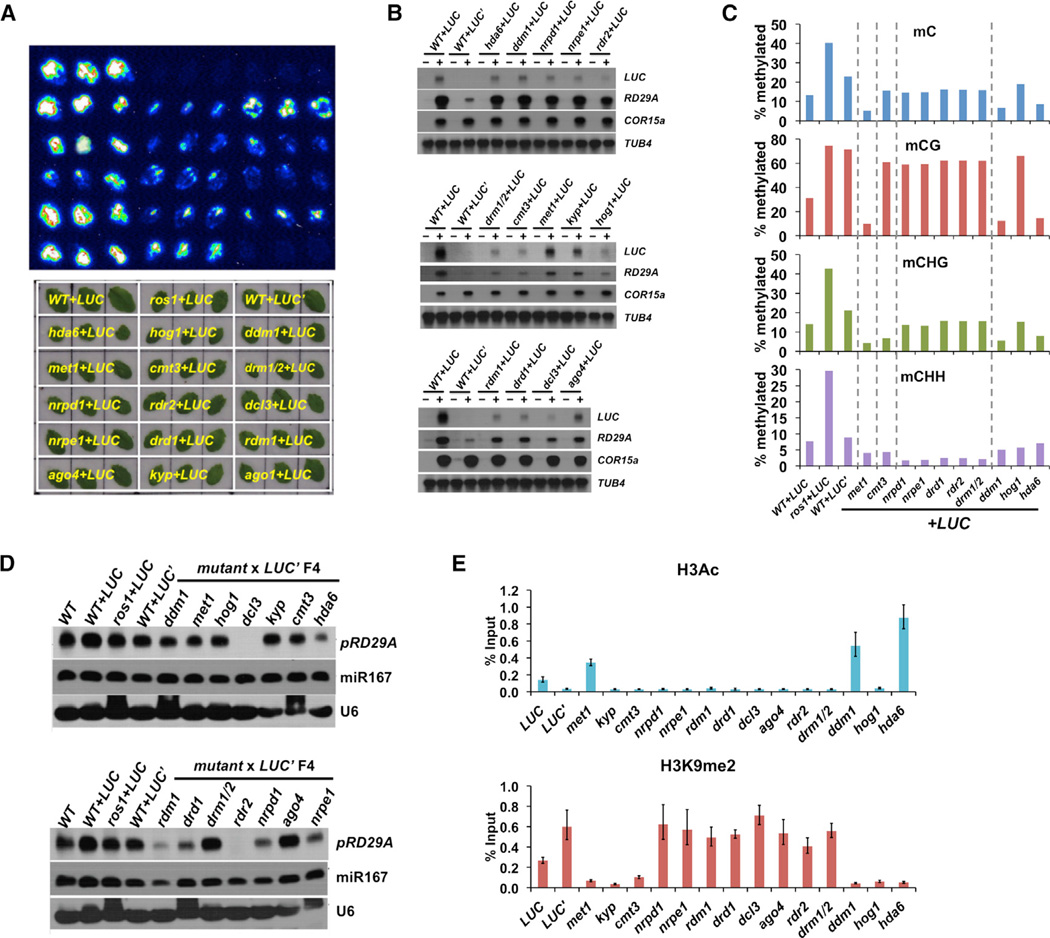
Figure 3. Multiple Epigenetic Pathways Are Required to Maintain LUC’ Silencing.
(A) Bioluminescence and bright field imaging results using rosette leaves from mutant+LUC plants. Genotypes of the plants are marked in yellow on the bright field image. The F3 plants used for the analyses were pre-screened for the presence of pRD29A-LUC transgene.
(B) Transcript levels of pRD29A-LUC and endoRD29A genes in the F3 mutant plants are examined by northern blotting. Please note that the signals from WT+LUC plants differ on different blots due to different exposure time, which serve as a positive control. Stress-treated and control plants are indicated by − and +, respectively. TUB4 and COR15A each serve as the loading control and the control for normal cold response.
(C) DNA methylation levels at the transgenic RD29A promoter in the mutant+LUC’ crosses F4 plants as measured by bisulfite sequencing.
(D) Northern blotting analyses of 24-nt siRNAs generated from the RD29A promoter (endogenous + transgenic). U6 snoRNA and miR167 each serves as the loading control and microRNA pathway control.
(E) Chromatin immunoprecipitation followed by quantitative PCR was used to examine histone H3 acetylation and H3K9me2 levels at the transgenic RD29A promoter. Error bars indicate SD calculated from qPCR reactions of three technical replicates.