Figure 5.
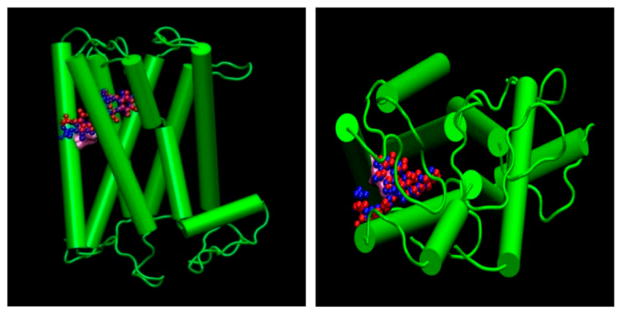
Side ((A) on left) and top ((B) on right) view of the known agonist docked to the rhodopsin-based homology model. Agonist 3D geometry optimization has been performed by Gaussian HF/6-31G* basis (Blue) and Marvin Chemaxon MM force field (Red) structure.
