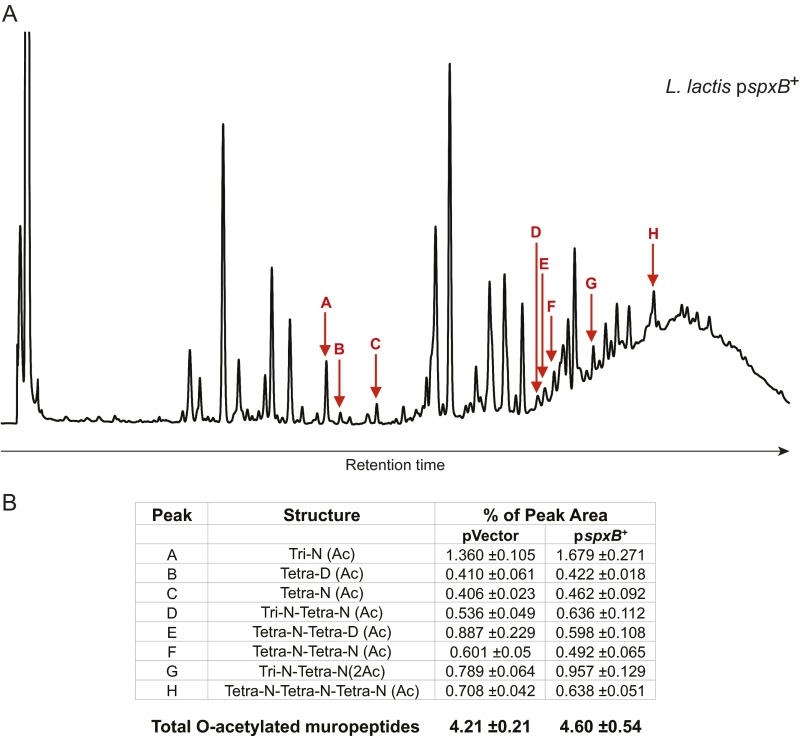
Fig. S3.
Peptidoglycan structure analysis of L. lactis pspxB+. (A) Representative HPLC profile of muropeptides obtained by mutanolysin digestion of peptidoglycan extracted from L. lactis pspxB+. Red arrows indicate O-acetylated muropeptides. (B) Table listing O-acetylated muropeptides and their respective percent abundances in L. lactis pspxB+ and the plasmid vector control strain. Ac, acetylation; D, d-Asp; Disaccharide, GlcNAc-MurNAc; iGln, α-amidated isoGlu (or γGlu); N, d-Asn; Tetra, disaccharide tetrapeptide (l-Ala-d-iGln-l-Lys-d-Ala); Tri, disaccharide tripeptide (l-Ala-d-iGln-l-Lys). The total percentage of O-acetylated muropeptides was calculated as follows: percentage = Σ monomers (Ac) + 1/2 Σ dimers (Ac) + Σ dimers (2Ac) + 1/3 Σ trimers (Ac). Data represent mean ± SD of three independent culture and media preparations.