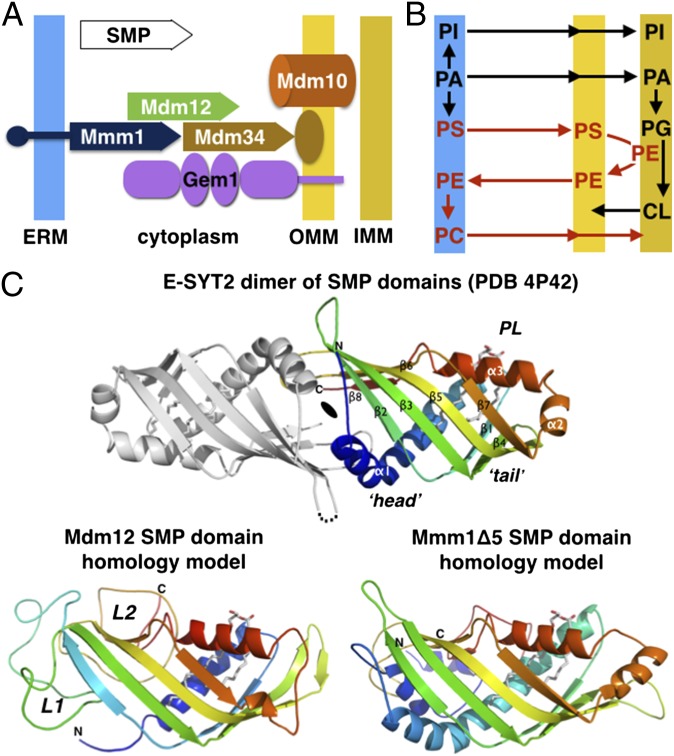
Fig. 1.
ERMES, phospholipid metabolism, and the signature SMP domain. (A) Subunit topology and composition of the ERMES complex. The three SMP domain-containing subunits Mdm12, Mmm1, and Mdm34 together with a β-barrel outer mitochondrial membrane protein Mdm10 and the calcium-activated regulatory GTPase Gem1 constitute the ERMES in yeast. The SMP domains are depicted with a plain arrow. (B) Interconnection of the phospholipid metabolic pathways at the interface between ER and mitochondria. (C, Upper) Homology models for the SMP domains of yeast Mdm12 and Mmm1. The crystal structure of the head-to-head dimer of SMP domain of E-SYT2 (23) present in an ER-to-plasma membrane tether is shown with the observed bound phospholipid (PL). Only one SMP monomer has been colored. (Lower) Phyre2 homology models of the yeast Mdm12 and Mmm1 SMP. L1 and L2 refer to the nonconserved insertions present in Mdm12 sequences.