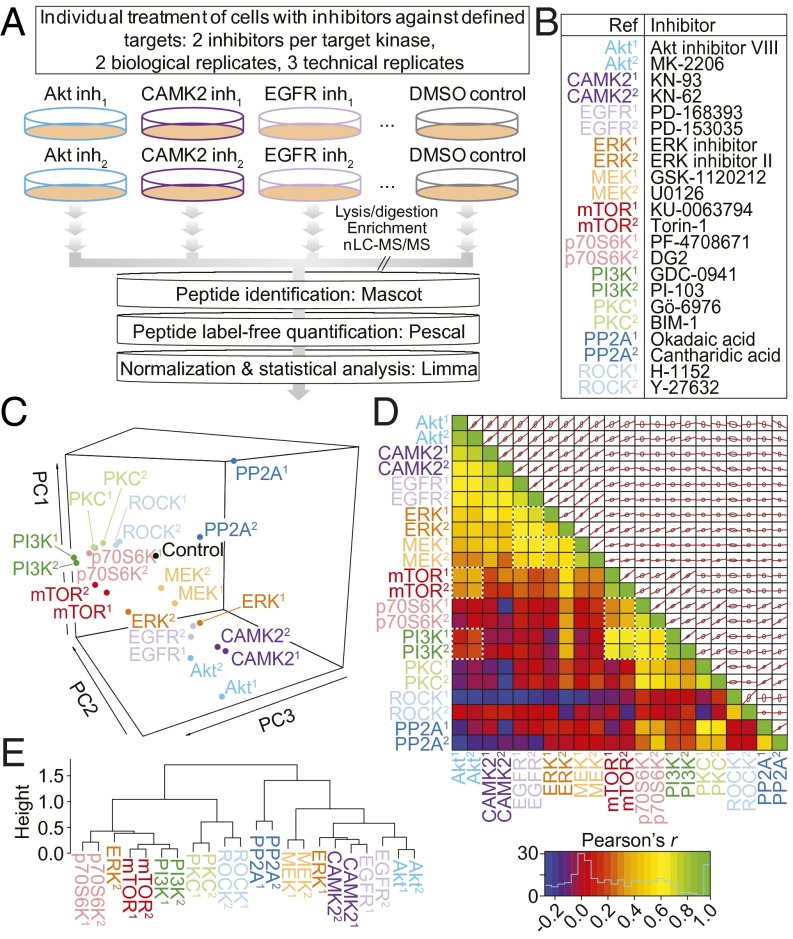
Fig. 1.
Phosphoproteomics data elucidate the relationships between kinase inhibitors. (A) Experimental design. Inh, inhibitor. (B) Kinase inhibitors used in the study. (C) PCA of the 4,651 phosphorylation sites whose abundance was reduced significantly (adjusted P < 0.05) by at least one inhibitor treatment. PC, principal component. (D) Lower triangle, Pearson correlation coefficients between each of the inhibitor treatments. Known kinase–kinase relationships are highlighted with white, dashed boxes. Upper triangle, pair-wise alignments of the 4,651 phosphorylation site log2 fold ratios for each inhibitor combination. Red lines indicate the linear model formed between the two variables; gray ellipses represent one SD from the mean in both dimensions. (E) Unsupervised, hierarchical clustering (Pearson correlation distance metric) of the mean log2 ratios for peptides containing common phosphorylation motifs represented in the filtered 4,651 phosphorylation sites.