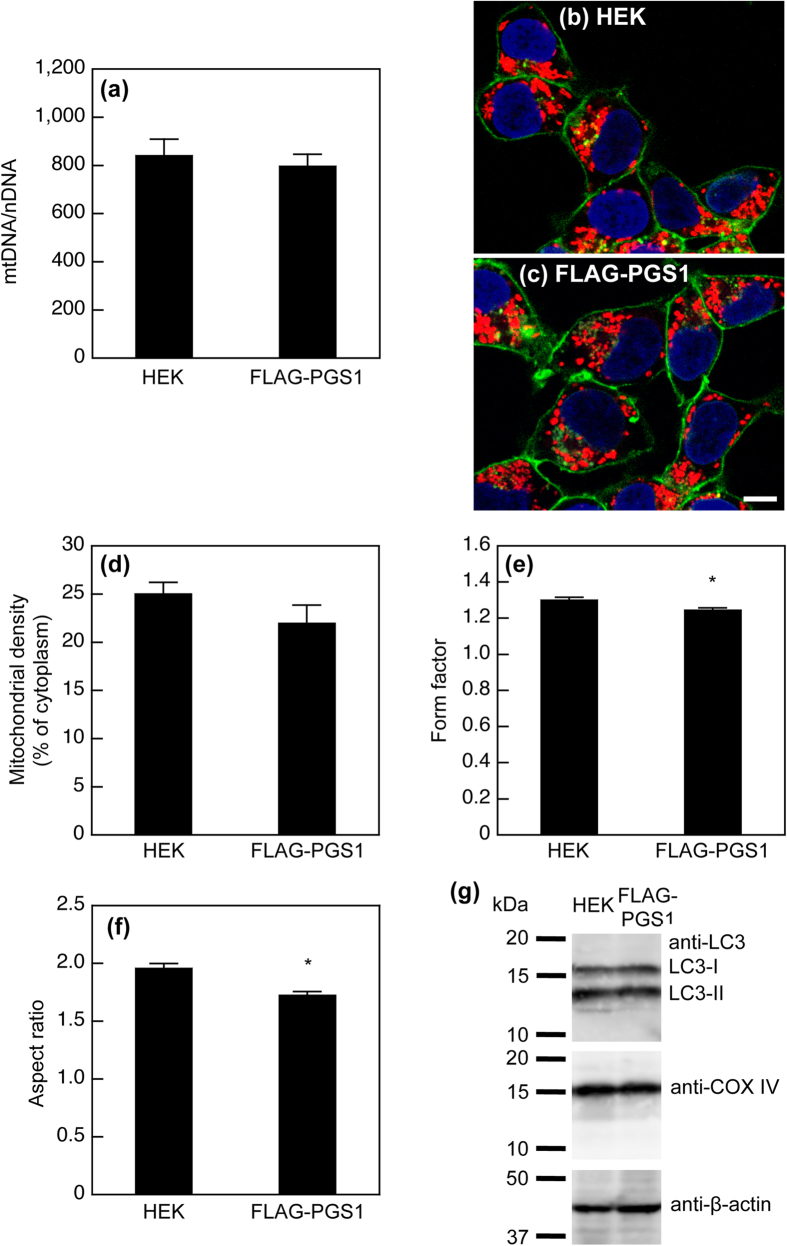
Figure 7. Effect of PGS1 overexpression on mitochondrial content and morphology.
(a) Total DNA was extracted from the cells, and the nuclear gene ASPOLG and the mitochondrial gene CCOI were quantified by real-time PCR. The results are expressed as the ratio of the mitochondrial DNA copy number to the nuclear DNA copy number (mtDNA/nDNA). Each bar represents the mean ± S.E. of six measurements. There was no statistically significant difference between HEK293 and HEK/FLAG-PGS1 cells. (b) and (c) Confocal fluorescence microscopy of HEK293 and HEK/FLAG-PGS1 cells. Mitochondria, nuclei, and cell surfaces were visualized with MitoTracker Red CMXRos (red), DAPI (blue), and FITC-concanavalin A (green), respectively. The bar represents 20 μm. Confocal images were used to determine mitochondrial densities, form factors and aspect ratios. (d) Mitochondrial density is expressed as the percentage of cytoplasmic area occupied by mitochondria. Each bar represents the mean ± S.E. from 18 cells. (e) and (f) The form factor (perimeter2/4π·area) and the aspect ratio (major axis/minor axis) were calculated for each mitochondrial object (n = 427, HEK293; n = 410, HEK/FLAG-PGS1) in 18 cells per group. Both parameters have a minimal value of 1, which represents a perfect circle, and the values increase as the shape becomes elongate. Each bar represents the mean ± S.E. *P < 0.05, significantly different from HEK293 cells. (g) Immunoblot analysis of LC3. Sonically disrupted cell lysates (37.0 μg of protein) from HEK293 and HEK/FLAG-PGS1 cells were separated by 15% SDS-PAGE. LC3, COX IV, and β-actin were detected with specific antibodies.