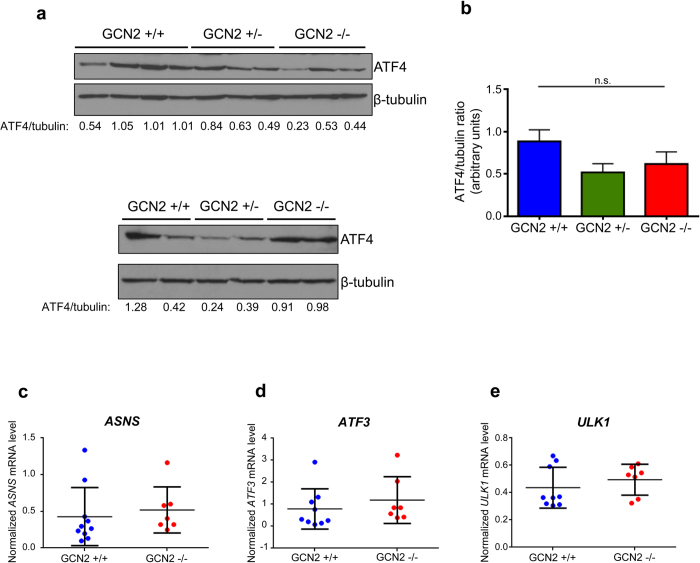
Figure 5. GCN2−/− mixed background sarcomas do not express lower levels of ATF4, in spite of reduced eIF2α phosphorylation.
(a) Western blot analysis on two independent samples of tumor homogenates for levels of ATF4 in mixed background tumors. β-tubulin was used as a loading control. The ratio of ATF4 to β-tubulin is displayed below each blot. (b) Graphical representation of the ATF4 to β-tubulin ratios calculated from the blots in (a). Data are represented as the average value for each genotype ± standard deviation. Results are not statistically significant. (c) qPCR analysis of ASNS levels in GCN2+/+ and GCN2−/− sarcomas. ASNS levels were normalized to the geometric mean of the reference genes β-tubulin, β-actin, and 18S rRNA. Data are represented as the average value for each genotype ± standard deviation. Results are not statistically significant. (d) qPCR analysis of ATF3 levels in GCN2+/+ and GCN2−/− sarcomas. ATF3 levels were normalized to the geometric mean of the reference genes β-tubulin, β-actin, and 18S rRNA. Data are represented as the average value for each genotype ± standard deviation. Results are not statistically significant. (e) qPCR analysis of ULK1 levels in GCN2+/+ and GCN2−/− sarcomas. ULK1 levels were normalized to the geometric mean of the reference genes β-tubulin, β-actin, and 18S rRNA. Data are represented as the average value for each genotype ± standard deviation. Results are not statistically significant.