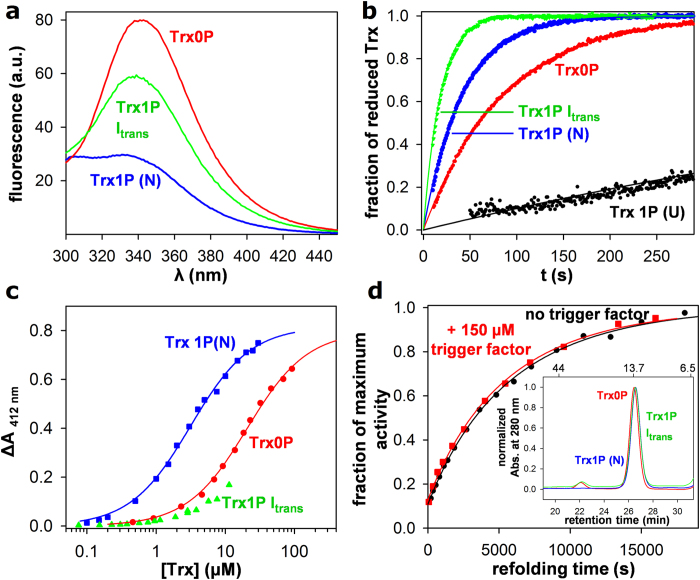
Figure 6. Biophysical and functional properties of Trx1P Itrans compared to Trx0P and native Trx1P at 0.2 M GdmCl, 25 °C and pH 7.0.
a: Fluorescence spectra of Trx0P, native Trx1P and Trx1P Itrans. b: Reduction of Trx0P, Trx1P, and Trx1P Itrans (2 μM each; red, blue and green dots, respectively) by 20 μM DTT, fitted according to second-order kinetics (solid lines) (see Supplementary Table 3 for the deduced rate constants and errors). Reduction of unfolded Trx1P with 100 μM DTT (black dots) revealed a rate constant of 10.3 ± 1.48 M−1 s−1) and was thus more than 100-fold slower than reduction of native Trx1P or Itrans (cf. Supplementary Table 3). c: Kinetic analysis documenting the properties of Trx0P (red circles), Trx1P (blue squares) and Trx1P Itrans (green triangles) as substrate of TrxR in the presence of 20 mM GdmCl at pH 8.0. Similar to Trx0P, Itrans showed an about 10-fold increase in KM relative to that of Trx1P, but its catalytic parameters could not be determined accurately due to unspecific aggregation at higher concentrations (cf. Supplementary Table 3 for the parameters and errors deduced from Michaelis Menten fits). d: Itrans has a compact tertiary structure with a buried trans Ile75-Pro76 peptide bond that is inaccessible to catalysis by the PPIase trigger factor. Formation of native Trx1P from Itrans (90 μM) in the absence (black circles) and presence (red squares) of excess trigger factor (150 μM) was monitored by the increase in TrxR substrate activity and fitted monoexponentially (solid lines). Inset: Size exclusion chromatography on Superdex 75 of Itrans, native Trx1P and native Trx0P (oxidized forms), showing that Itrans has same hydrodynamic volume as native Trx1P. The retention time of all proteins at ~26.5 min is about ¼ of the half-life of Itrans folding (99 min), guaranteeing that only a minor fraction of Itrans reacted to N during the chromatography. Retention times of molecular mass standard proteins (in kDa) are indicated on the top.