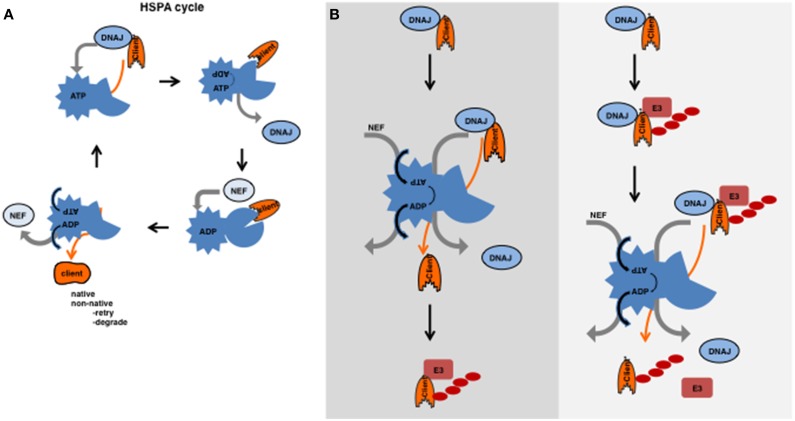
Figure 1.
(A) HSPA cycle. HSPAs (HSP70s and HSC70s) contain an N-terminal ATPase domain connected by a hydrophobic linker to a variable C-terminal peptide-binding domain (Hartl et al., 2011). This peptide-binding domain binds to a stretch of hydrophobic residues flanked by positive residues, which is predicted to occur every 40 amino acids (Frydman et al., 1999). In the folded state, these hydrophobic residues are buried inside, but are exposed in the unfolded or misfolded state. The energy obtained from the hydrolysis of ATP is required for assisted folding. However, the exact mechanism how an HSPA supports folding is still not yet completely understood. HSPA functions with the help of co-chaperones that orchestrate the cycle of ATP hydrolysis and substrate/client binding and release. DNAJs recognize clients and subsequently bind to the ATP-bound form of HSPA; upon binding of the DNAJ-client complex, ATP is hydrolysed by the HSPA and the DNAJ is released. Upon hydrolysis and DNAJ release, the peptide-binding-domain of HSPA undergoes a conformational change and clams around the polypeptide (the substrate) (Jiang et al., 2007; Swain et al., 2007; Bertelsen et al., 2009). The NEF has affinity to the ADP-bound form and mediates the exchange of ADP for ATP. The client has less affinity for the ATP-bound form of HSPA and releases together with the NEF. As a result, the client can fold or will re-enter the cycle if not completely folded, or somehow can be transferred to degradation machineries. (B) Substrates ubiquitylated before or after the action of HSPA. On the left, the canonical model is depicted in which an intrinsically unstable substrate is first recognized by a DNAJ then transferred to the HSPA cycle and after a futile folding event ubiquitylated and subsequently degraded. On the right, a model is presented in which an unfolded substrate is recognized by a DNAJ after which an E3 ligase can ubiquitylate it and the HSPA cycle acts on the ubiquitylated substrate. After the action of HSPA, the substrate is liberated from both the DNAJ and the E3 ligase (complex) and targeted for degradation.