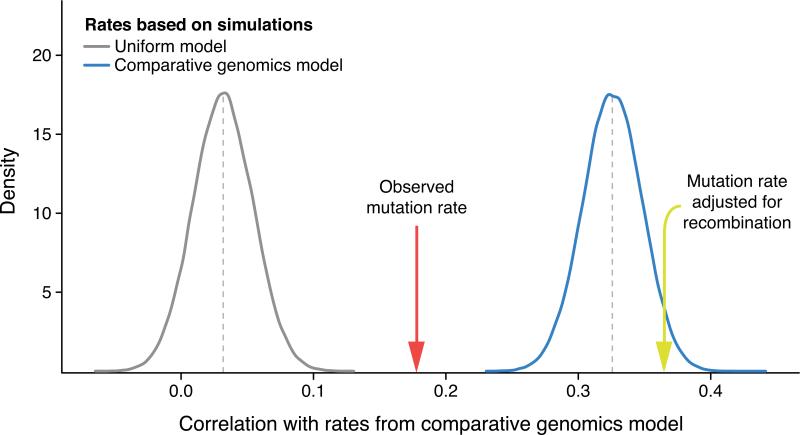
Fig. 4. Influence of mutation and recombination rates on human-chimpanzee divergence.
The correlation to substitution rates computed from a human-chimpanzee comparative genomics (HCCG) model is plotted for mutation rates inferred from a uniform mutation rate model (grey distribution) and for mutation rates inferred from the HCCG model itself (blue distribution), both based on 100,000 simulations of N = 11,020 mutations and binned in 1Mb windows. By sampling the same number of mutations, comparisons between rate estimates are meaningful. The effect of sampling is illustrated by the mean correlation of the HCCG with itself at only 0.33, which would asymptotically reach unity with infinite sampling. The correlation is also given for observed de novo mutation rates (red arrow, N = 11,020) and observed de novo mutation rates adjusted for local recombination rates 31 (yellow arrow, N = 11,020). Correlation with observed de novo mutation rates (r = 0.18) is stronger than correlations with rates based on the uniform model (mean r = 0.032, p < 1 × 10−5), indicating that the HCCG model partly captures regional mutation rate variations. However, the correlation with observed de novo mutation rates is weaker than correlations with rates based on the HCCG model itself (mean r = 0.33, p < 1 × 10−5), suggesting other contributing factors. When adjusting observed de novo mutation rates for local recombination rates, the correlation is 0.37, illustrating that substitution rates computed from the HCCG model capture both mutation rates and orthogonal evolutionary forces associated with local recombination rates.