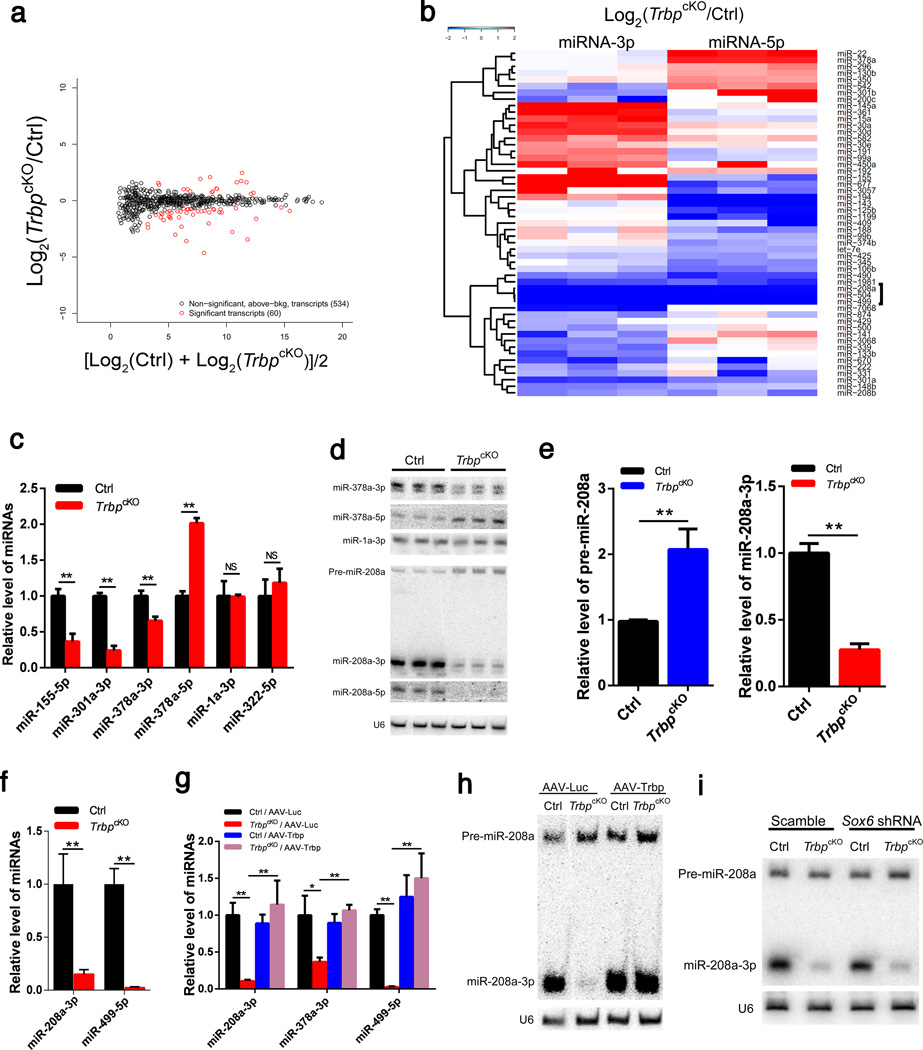
Figure 5. Genome-wide identification of dysregulated miRNA species in TrbpcKO hearts.
a) Scatter plot of the expression of 594 miRNAs between p2.5 TrbpcKO and control hearts. 60 dysregulated miRNA species (derived from 53 miRNA genes) are marked red.
b) Hierarchical clustering heatmap of the 53 dysregulated miRNA genes between TrbpcKO (n = 3) and control (n = 3) hearts. Significantly down-regulated miR-208a cluster is marked.
c) qRT-PCR of indicated miRNAs in p2.5 TrbpcKO and control hearts. n = 3.
d) Northern blot of indicated miRNAs in p2.5 TrbpcKO and control hearts. U6 serves as a loading control.
e) Quantification of pre-miR-208a (left panel) and mature miR-208a (right panel) from Northern blots of p2.5 TrbpcKO and control hearts. n = 3.
f) qRT-PCR of miR-208a-3p and miR-499-5p in p2.5 TrbpcKO and control hearts. n = 3.
g) qRT-PCR of miR-208a-3p, miR-378a-5p and miR-499-5p in the hearts of 1-month old TrbpcKO and control mice injected with AAV-Trbp or AAV-Luc control. n = 3.
h) Northern blot of miR-208a-3p in the hearts of 1-month old TrbpcKO and control mice injected with AAV-Trbp or AAV-Luc control. Both the precursor (pre-miR-208a) and mature product (miR-208a-3p) are shown. U6 serves as a loading control.
i) Northern blot of miR-208-3p in the hearts of 1-month old TrbpcKO and control mice injected with AAV-Scramble or AAV-Sox6 shRNA. Both the precursor (pre-miR-208a) and mature product (miR-208a-3p) are shown. U6 serves as a loading control. Values are expressed as the mean ± SD. NS: not significant, *: P<0.05, **: P<0.01.