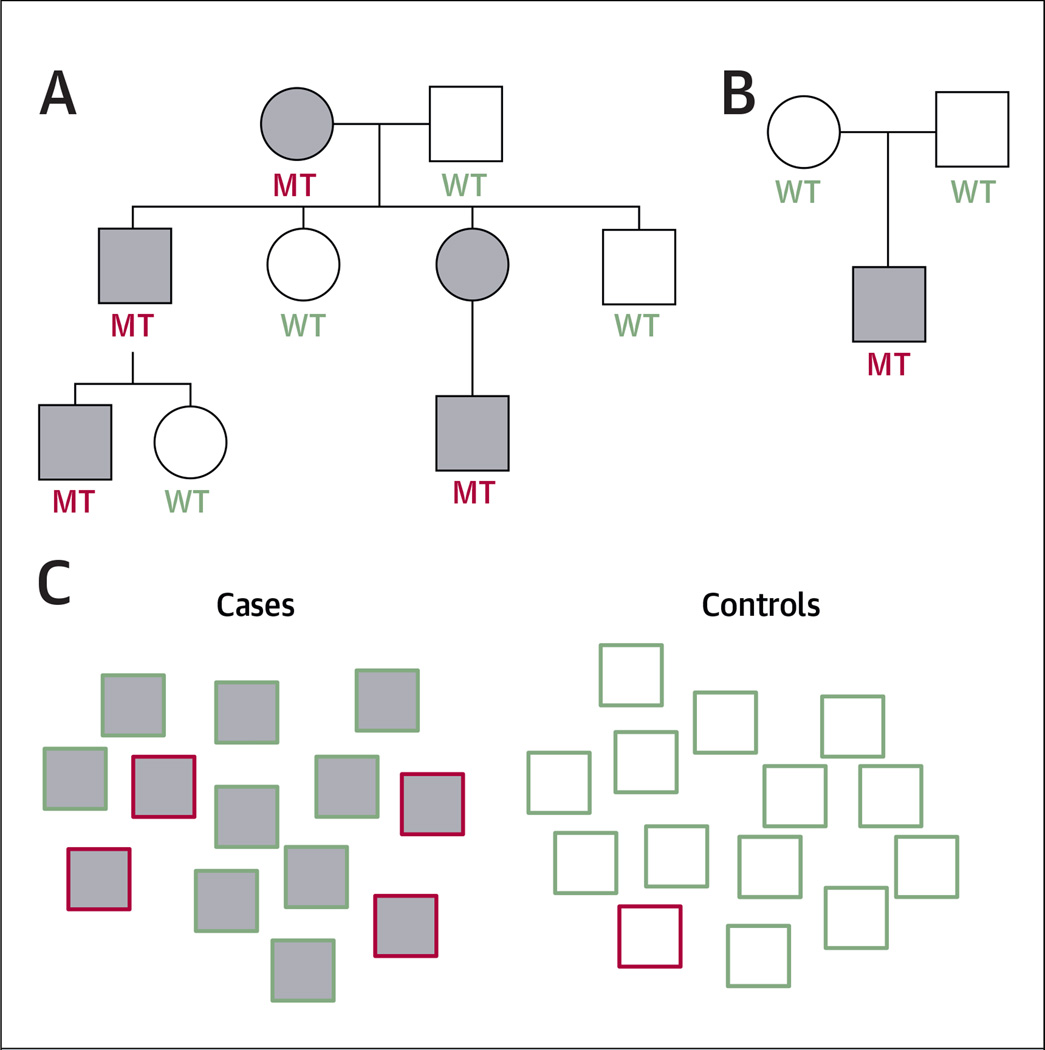
FIGURE 2. Experimental Designs to Identify Disease-Causing and Susceptibility Genetic Variants.
(A) Example of a 3-generation pedigree with autosomal-dominant inheritance. Affected patients are gray. Mutation carriers (MT) and subjects without mutations (WT) are highlighted in red and green, respectively. In this instance, the penetrance of the mutation is complete, and the mutant gene can be mapped to a segment of the genome using linkage analysis. (B) Example of a trio. De novo mutations in the affected offspring that are not found in the parents may be prioritized for analysis in independent groups of cases. In this situation, the penetrance of the phenotype in the parents is uncertain and may confound the results. (C) Example of a case-control association study. The genotypes of large groups of cases (gray) and controls (white) are compared at sites across the genome. The red border identifies subjects who carry the risk allele that is being tested for association. Variants that are enriched in cases compared with controls are considered to be associated with the disease. In this case, there is a 4-fold enrichment of the variant among the cases compared with controls.