Figure 7.
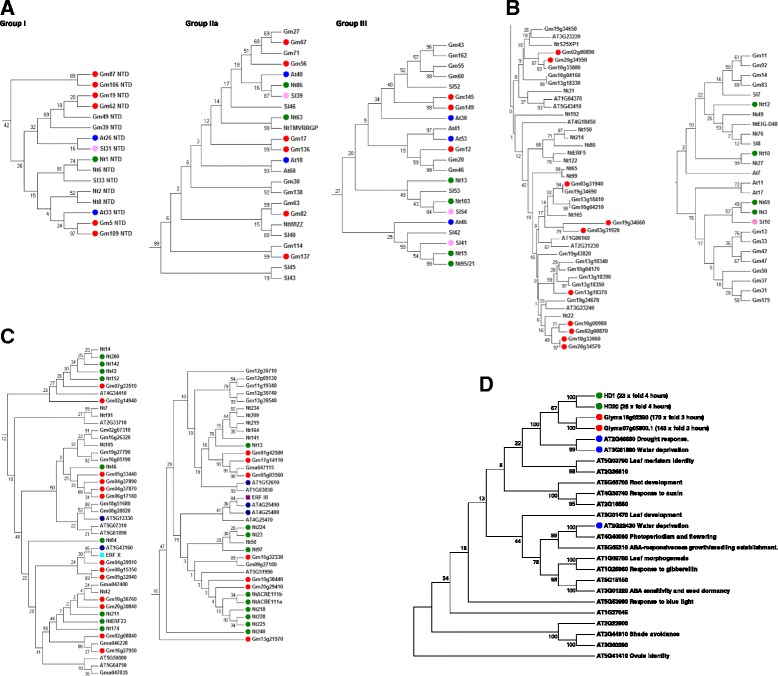
Drought-inducible transcription factor genes in the WRKY, HDZIP, and AP2/ERF families. A. Core responses in the WRKY transcription factor family B. Family/species-specific responses in the AP2/ERF and WRKY families C. Core responses in the AP2/ERF family D. Core responses in the HDZIP family. Drought stress inducibility or association with the GO term “Response to water deprivation” (GO:0009414) is denoted by red circles (soybean), blue circles (Arabidopsis), green circles (tobacco), and pink circles (tomato). Marker domains for Group III and X ERF subfamilies are shown as purple and light blue squares respectively. The evolutionary histories were inferred using the Neighbor-Joining method. The optimal trees are shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. For the HDZIP transcription factors, induction by drought stress and the processes that the transcription factors are involved in are shown. AP2/ERF and homeodomain genes are shown by gene model name except for tobacco where the TOBFAC nomenclature is used. WRKY genes from Arabidopsis are shown by their commonly used names, soybean from our own analysis (Additional file 9: Table S6), tobacco genes were taken from TOBFAC and tomato genes from published sources.
