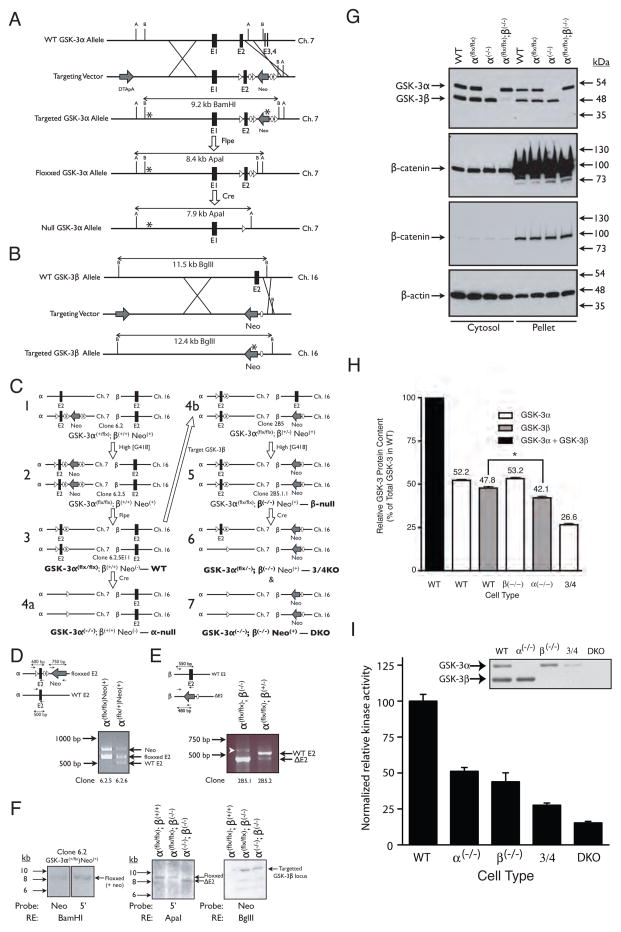
Figure 1.
Strategy used to create compound knockouts of GSK-3α and GSK-3β in ES cells. (A) Targeting strategy for GSK-3α. After homologous recombination with the targeting vector, exon 2 was replaced with a LoxP-flanked (floxxed) exon 2 and FRT-flanked (flrted) neomycin resistance cassette. B = BamHI; A = ApaI. (B) Targeting strategy for GSK-3β. After homologous recombination, exon 2 was replaced with a neomycin resistance cassette and a single FRT site. B = BglII. (C) Overview of the steps required to generate the various ESC lines used in this study (bold type). (D) Multiplex PCR analysis used to screen ESC clones for G418-mediated conversion from GSK-3α(+/flx) to GSK-3α (flx/flx) genotype (step 1–2 in panel C). (E) PCR analysis used to screen ESC clones for G418-mediated conversion from GSK-3α (flx/flx);GSK-3β(+/−) to GSK-3α(flx/flx);GSK-3β(−/−) genotype (step 4b-5 in panel C). White arrowhead indicates a non-specific band. In A-E, ovals represent FRT recombination sites, triangles represent LoxP recombination sites and asterisks represent hybridization sites for Southern blot probes. (F) Southern blots confirming proper targeting of GSK-3 loci. (G) Wild-type, GSK-3α(flx/flx), GSK-3α (−/−) and GSK-3β(−/−) ES cells all display the same level of cytosolic and membrane-associated (pellet) β–catenin. Two different exposures of the same β-catenin immunoblots are presented. (H) Quantitation of GSK-3 in WT and knockout cell lines. Three lysates, each from a different plate of cells, were analyzed per cell type. Error bars = SEM. * p <0.05 (ANOVA with Tukey’s post-hoc test). (I) Consequence of reduced GSK-3 gene dosage on kinase activity. Each bar represents the average from 2 independent kinase assay experiments with each assay performed in triplicate. Error bars = SEM. Inset – immunoblot of the GSK-3 band profiles in the eluates used in the kinase assays (normalized to total protein content).