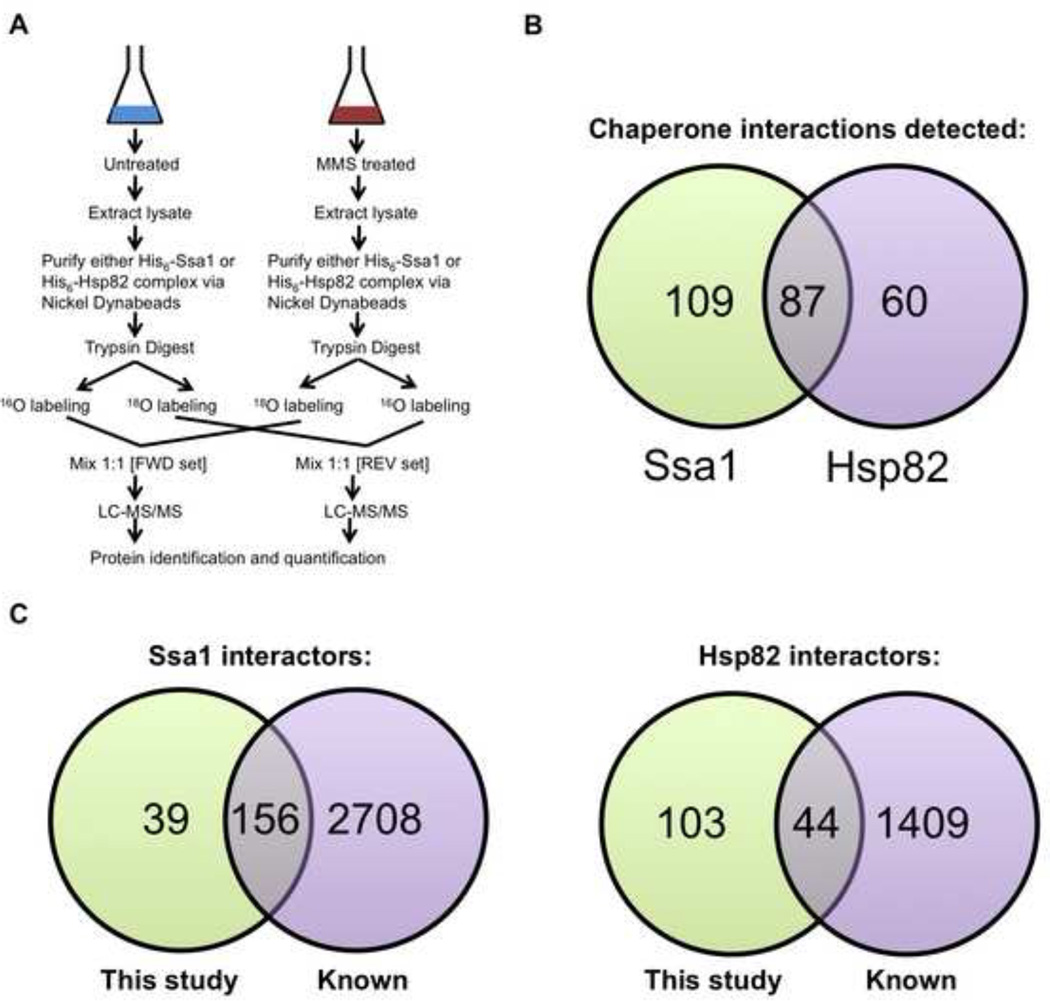
Figure 1. Quantitative affinity-purification mass spectrometry strategy for analysis of dynamic chaperone interactomes during DNA damage response.
(A) Growing yeast cells expressing His6-tagged Ssa1 or Hsp82 were left untreated or exposed to 0.02% methyl methanesulfonate (MMS) for 3 h. Chaperone interactomes were isolated by nickel-NTA magnetic bead affinity purification. Following PAGE and in-gel trypsin digestion and peptides were 18O or 16O labeled by trypsin-mediated exchange. Then, the heavy and light samples were combined and analyzed by Orbitrap LC-MS/MS and MassQuant informatic analysis, allowing identification of interacting proteins and determination of their relative enrichment after DNA damage. Each experiment was performed in biological triplicate from which each pair of samples was analyzed as technical replicates by forward and reverse 18O labeling.
(B) Venn diagram of candidate yeast Ssa1 and Hsp82 interactors remaining after applying statistical filters.
(C) Comparison of candidate yeast Ssa1 and Hsp82 interactors detected in our study vs. previously identified interactors in public databases.