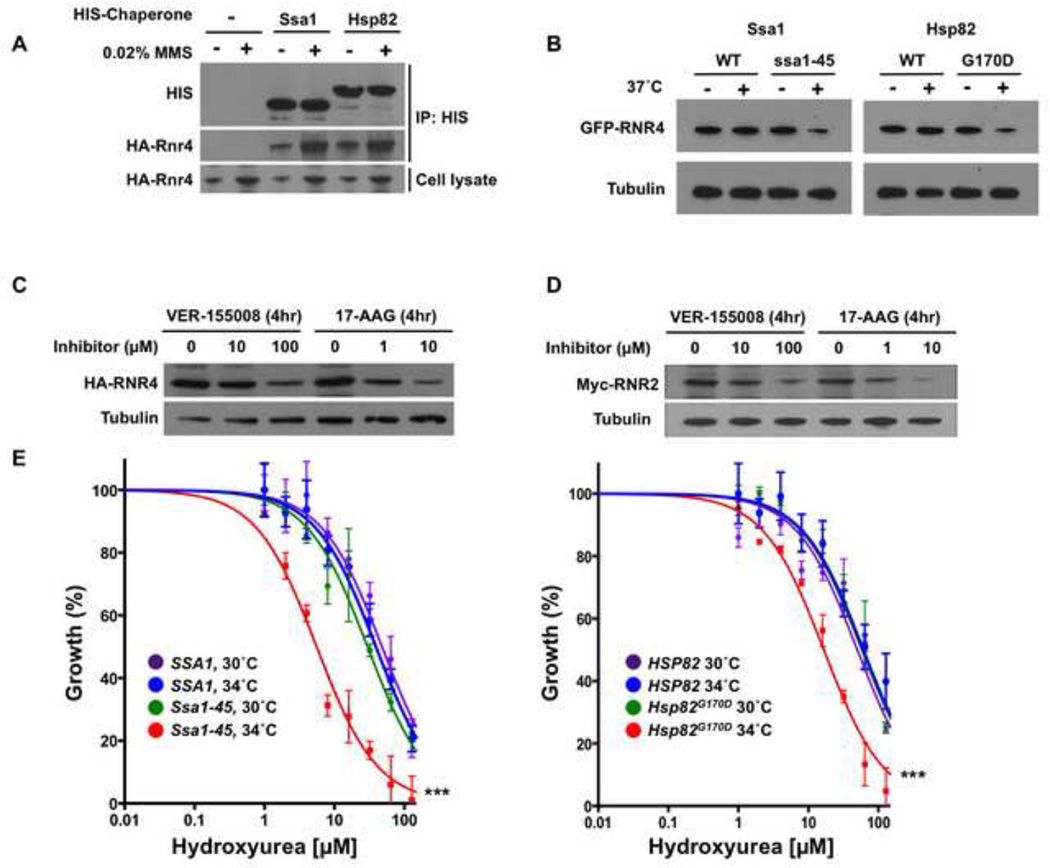
Figure 6. Ribonucleotide reductase is a client of Hsp70 and Hsp90 in yeast cells.
A) Yeast expressing HA-tagged Rnr4 and either untagged chaperones, HIS6-Ssa1 or HIS6-Hsp82 were grown to early mid-log phase. Cells were lysed, pull-down was performed with His-Tag Dynabeads, and captured proteins were separated by SDS-PAGE, blotted and probed with anti-hexahistidine or anti-HA antibody.
B) Yeast expressing GFP-tagged Rnr4 along with wild-type chaperones or temperature-sensitive alleles Ssa1-45 or Hsp82G170D were incubated at 25° or 37° C for 90 min. GFP-Rnr4 levels were determined via Western blot of cell lysates with anti-GFP antibody. Equal loading was determined using anti-tubulin immunoreactivity.
C) Yeast cells expressing HA-tagged Rnr4 were grown to early mid-log phase and treated with DMSO, Hsp70 inhibitor VER-155008 or Hsp90 inhibitor 17-AAG for 4 h. Cells were lysed and HA-Rnr4 levels were determined via Western blot with anti-HA epitope antibody and anti-tubulin loading control.
D) Yeast cells expressing Myc-tagged Rnr2 were grown to early mid-log phase and treated with DMSO, Hsp70 inhibitor VER-155008 or Hsp90 inhibitor 17-AAG for 4 h. Cells were lysed and Myc-Rnr2 levels were determined via Western blot with anti-Myc epitope antibody and anti-tubulin loading control.
E) Yeast expressing wild-type chaperones or temperature-sensitive alleles Ssa1-45 or Hsp82G170D were diluted to OD600 of 0.01 and grown for 8 h in rich media at either 25° or 34° C in the presence of hydroxyurea (HU) at the indicated concentrations. OD600 was determined, normalized to 0 µM HU (100%), the data for each condition fit to a nonlinear curve (Prism software) and plotted. Data shown are mean ± SEM of three replicates (*** P<0.001) compared to WT strain at 34° C, t-test)