Figure 2.
Processes Enriched in the ADP-Ribosylated Interactome
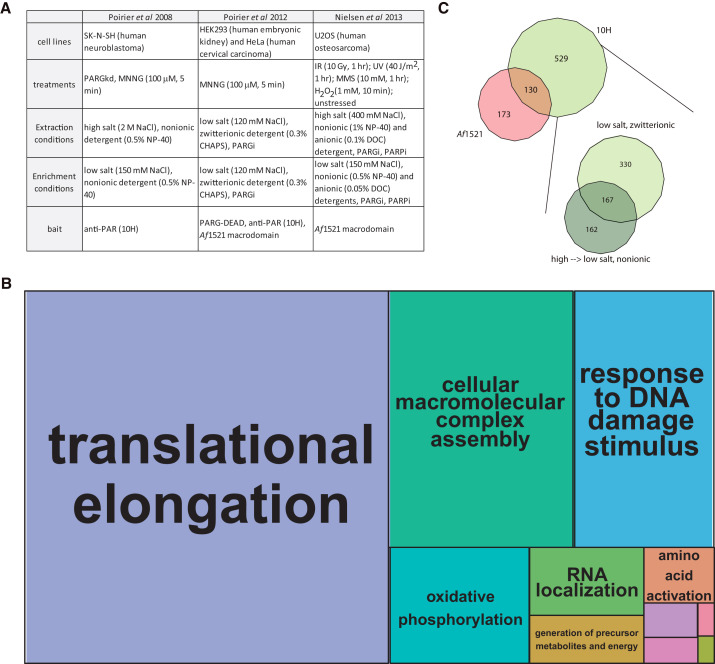
(A) Experimental design for the interactome studies used for this meta-analysis. PARGi, PARG inhibitor; PARPi, PARP inhibitor; PARGkd, PARG knockdown.
(B) The pooled DNA-damaged induced ADP-ribosylated interactome depicted as a treemap of enriched biological processes. The most enriched biological processes (based on statistical likelihood) are shown as larger components within the map and grouped according to common cellular functions. See Figure S1 for the detailed version of this treemap. Gene ontology determined using DAVID (Huang et al., 2009), treemap constructed using REViGO (Supek et al., 2011) and R (R Development Core Team, 2011).
(C) A compilation of the proteins identified in response to DNA damage can be broken out by enrichment methods (bait) or cell lysis conditions. For comparison of lysis conditions, the 10H enriched proteins were analyzed. Euler diagrams created in VennMaster (Kestler et al., 2005). Source data available in Table S2.