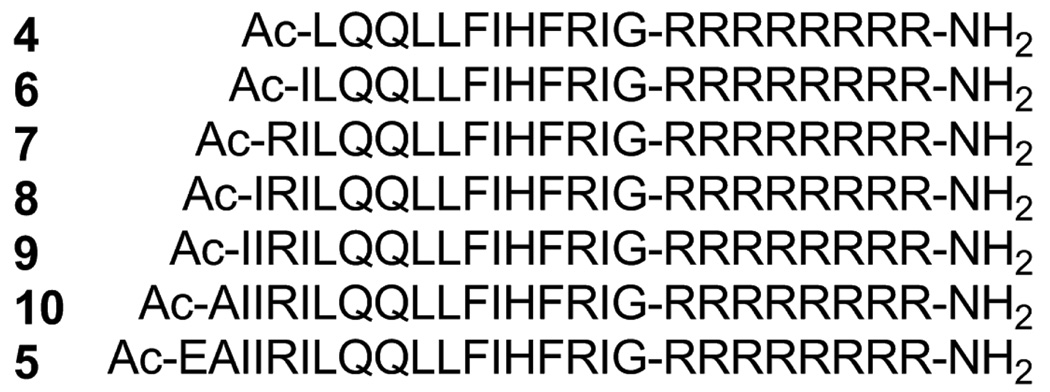Abstract
Structure-activity relationship studies were conducted on HIV integrase (IN) inhibitory peptides which were found by the screening of an overlapping peptide library derived from HIV-1 gene products. Since these peptides located in the second helix of Vpr are considered to have an alpha-helical conformation, Glu-Lys pairs were introduced into the i and i+4 positions to increase the helicity of the lead compound possessing an octa-arginyl group. Ala-scan was also performed on the lead compound for the identification of the amino acid residues responsible for the inhibitory activity. The results indicated the importance of an alpha-helical structure for the expression of inhibitory activity, and presented a binding model of integrase and the lead compound.
1. Introduction
Highly active anti-retroviral therapy (HAART), which involves a combination of two or three agents from two categories, reverse transcriptase inhibitors and protease inhibitors, has brought us remarkable success in the clinical treatment of HIV-infected and AIDS patients.1 However, it has been accompanied by serious clinical problems including the emergence of viral strains with multi-drug resistance (MDR), considerable adverse effects and nonetheless high costs. As a result, new categories of anti-HIV agents operating with mechanisms of action different from those of the above inhibitors are sought. HIV-1 integrase (IN) is a critical enzyme for the stable infection of host cells since it catalyzes the insertion of viral DNA into the genome of host cells, by means of strand transfer and 3’-end processing reactions and thus it is an attractive target for the development of anti-HIV agents. Recently, the first IN inhibitor, raltegravir (Merck),2 has appeared in a clinical setting. It is assumed that the activity of IN must be negatively regulated during the translocation of the viral DNA from the cytoplasm to the nucleus to prevent auto-integration. The virus, as well as the host cells, must encode mechanism(s) to prevent auto-integration since the regulation of IN activity is critical for the virus to infect cells.3 By screening a library of overlapping peptides derived from HIV-1 SF2 gene products we have found three Vpr-derived peptides, 1, 2 and 3, which possess significant IN inhibitory activity, indicating that IN inhibitors exist in the viral pre-integration complex (PIC).4 The above inhibitory peptides, 1, 2 and 3, are consecutive overlapping peptides (Figure 1). Compounds 4 and 5 are 12- and 18-mers from the original Vpr sequence with the addition of an octa-arginyl group5 into the C-terminus for cell membrane permeability, respectively. Compounds 4 and 5 have IN inhibitory activity and anti-HIV activity. Here we report structure-activity relationship studies on these lead compounds for the development of more potent IN inhibitors.
Figure 1.
Amino acid sequences of compounds 1–5. Compounds 1–3 are consecutive overlapping peptides with free N-/C-terminus. These were found by the IN inhibitory screening of a peptide library derived from HIV-1 gene products. Compounds 4 and 5 are cell penetrative leads of IN inhibitors.
2. Results and discussion
To determine which lead compound is most suitable for further experiments, five peptides 6–10, which were elongated by one amino acid starting with compound 4 and extended ultimately to 5, were synthesized (Figure 2). Judging by the 3’-end processing and strand transfer reactions in vitro,6 these peptides 4–10 had similar inhibitory potencies (Table 1). As a result, we concluded that 12 amino acid residues derived from the original Vpr sequence are of sufficient for IN inhibitory activity, and any peptide among 4–10 is a suitable lead.
Figure 2.
Amino acid sequences of compounds 6–10, which are elongated by one amino acid from compound 4 to 5.
Table 1.
IC50 values of compounds 4–10 toward the 3’-end processing and strand transfer reactions catalyzed by HIV-1 IN.
| compound | IC50 (µM) | |
|---|---|---|
| 3’-end processing | strand transfer | |
| 4 | 0.13 ± 0.02 | 0.06 ± 0.01 |
| 5 | 0.09 ± 0.01 | 0.04 ± 0.01 |
| 6 | 0.10 ± 0.01 | 0.07 ± 0.01 |
| 7 | 0.13 ± 0.02 | 0.11 ± 0.01 |
| 8 | 0.26 ± 0.04 | 0.11 ± 0.03 |
| 9 | 0.11 ± 0.01 | 0.07 ± 0.01 |
| 10 | 0.08 ± 0.01 | 0.05 ± 0.01 |
Structural analysis showed that the Vpr-derived peptides, 1, 2 and 3, are located in the second helix of Vpr and were thus considered to have an α-helical conformation.7 Compound 5 was adopted as a lead for the development of compounds with an increase in α-helicity since a longer peptide is likely to form a more stable α-helical structure than a shorter one. Initially, Glu (E) and Lys (K) were introduced in pairs into compound 5 at the i and i+4 positions. In general, such disposition of Glu-Lys pairs at i and i+4 positions is considered to cause an increase in α-helicity due to formation of an ionic interaction of a β-carboxy group of Glu and an ε-amino group of Lys. Several analogs of 5 with Glu-Lys pairs were synthesized by Fmoc-solid phase peptide synthesis (Figure 3). In the inhibitory assay against the 3’-end processing and strand transfer reactions catalyzed by HIV-1 IN in vitro, compounds 11 and 15 showed more potent inhibitory activities than 5 (Table 2). Substitution of Glu-Lys for His14-Gly18 or Ile3-Leu7 caused no decrease in IN inhibitory activity but a significant increase in activity, suggesting that Ile3, Leu7, His14 and Gly18 are not indispensable for activity. Substitution of Glu-Lys for Ala2-Ile6 or Gln9-Ile13 caused a slight decrease in IN inhibitory activity against the 3’-end processing and strand transfer reactions (compounds 12 and 13), indicating that Ala2 and/or Ile6, and Gln9 and/or Ile13 are partly required for activity. Substitution of Glu-Lys for Ile4-Gln8 caused a 2–4 fold decrease in IN inhibitory activity against the 3’-end processing and strand transfer reactions (compound 14), showing that Ile4 and/or Gln8 are essential for activity. Substitution of Glu-Lys for Leu11-Phe15 caused an 8-fold decrease in IN inhibitory activity against the 3’-end processing reaction and a 1.5-fold decrease in IN inhibitory activity against the strand transfer reaction (compound 16), indicating that Leu11 and/or Phe15 are indispensable for activity, especially for inhibition against 3’-end processing. Compound 17 has two substitutions of Glu-Lys for His14-Gly18 and for Ala2-Ile6, which are common to compounds 11 and 12, respectively. A 2-fold decrease in both IN inhibitory activities of compound 17 is mostly due to the substitution for Ala2-Ile6 common to 12, although 17 is slightly less active than 12 in both IN inhibitory assays.
Figure 3.
Amino acid sequences of compounds 11–17, into which Glu-Lys pairs have been introduced.
Table 2.
IC50 values of compounds 5 and 11–17 toward the 3’-end processing and strand transfer reactions catalyzed by HIV-1 IN.
| compound | IC50 (µM) | |
|---|---|---|
| 3’-end processing | strand transfer | |
| 5 | 0.09 ± 0.01 | 0.04 ± 0.01 |
| 11 | 0.05 ± 0.01 | 0.01 ± 0.001 |
| 12 | 0.12 ± 0.01 | 0.047 ± 0.01 |
| 13 | 0.14 ± 0.02 | 0.065 ± 0.01 |
| 14 | 0.23 ± 0.03 | 0.15 ± 0.002 |
| 15 | 0.04 ± 0.01 | 0.031 ± 0.01 |
| 16 | 0.71 ± 0.21 | 0.06 ± 0.004 |
| 17 | 0.18 ± 0.06 | 0.08 ± 0.02 |
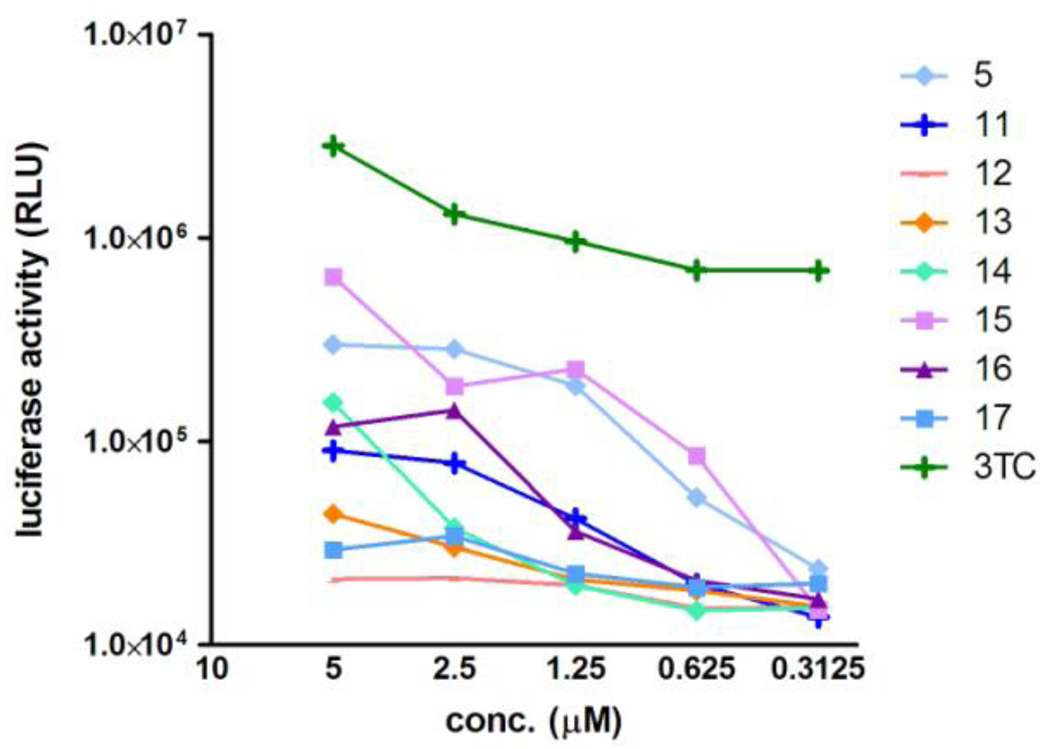
Anti-HIV activity of these compounds was assessed by an MT-4 Luc system, in which MT-4 cells were stably transduced with the firefly luciferase expression cassette by a murine leukemia viral vector. MT-4 Luc cells constitutively express high levels of luciferase. HIV-1 infection significantly reduces luciferase expression due to the high susceptibility of MT-4 cells to HIV-1 infection. Protection of MT-4 Luc cells from HIV-1-induced cell death maintains the luciferase signals at high levels. In addition, the cytotoxicity of test compounds can be evaluated by a decrease of luciferase signals in these MT-4 Luc systems. The parent compound 5 showed significant anti-HIV activity at concentrations above 1.25 µM, as reported previously (Figure 4).4 Compound 15 showed a significant inhibitory effect against HIV-1 replication, and is thus comparable to compound 5. Compounds 11, 14 and 16 also displayed weak antiviral effects at concentrations of 2.5 and 5.0 µM and compounds 12, 13 and 17 failed to show any significant anti-HIV activity. These results suggest that there is a positive correlation between IN inhibitory activity anti-HIV activity of the compounds. None of these compounds showed significant cytotoxic effects at concentrations below 5.0 µM.
Figure 4.
Luciferase signals in MT-4 Luc cells infected with HIV-1 in the presence of different concentrations of compounds 11–17. Luciferase activity is expressed as relative luciferase units (RLU). 3TC is an HIV reverse transcriptase inhibitor, which was used as a positive control.
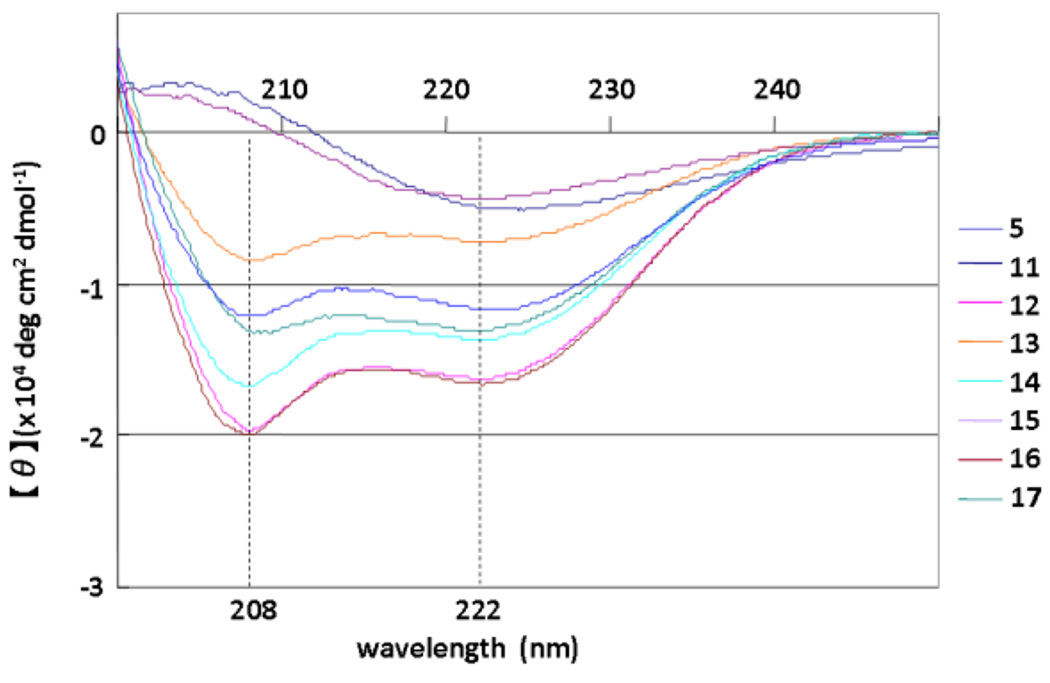
The structures of compounds 5 and 11–17 were assessed by CD spectroscopy. Because the aqueous solubility of these peptides is not high the peptides were dissolved in 0.1 M phosphate buffer, containing 50% MeOH at pH 5.6. The CD spectra suggest that the parent compound 5, which has no Glu-Lys pair, forms a typical α-helical structure, and the other compounds, with the exception of 11 and 15, form α-helical structures similarly (Figure 5). The order of strength of α-helicity is 12, 16 > 14 > 17 > 5 > 13. Compounds 11 and 15 have no characteristic pattern, although IN inhibitory activities of both compounds are superior to that of the parent compound 5. Replacement of His14-Gly18 and Ile3-Leu7 by Glu-Lys in compounds 11 and 15, respectively, caused a significant decrease in α-helicity, possibly due to formation of unfavorable salt bridges such as Glu14-Arg16 and Glu3-Arg5. Introduction of a Glu-Lys pair into Gln9-Ile13 in compound 13 caused a slight decrease in α-helicity, possibly due to interference in the formation of a salt bridge of Glu1-Arg5 by that of Arg5-Glu9. In the other analogs, increases in α-helicity were observed to result from the introduction of Glu-Lys pairs as we had initially postulated. Overall, there is no positive correlation between IN inhibitory or anti-HIV activity and the degree of α-helicity of the compounds.
Figure 5.
CD spectra of compounds 5 and 11–17 (5 µM) in 0.1 M phosphate buffer, pH 5.6 containing 50% MeOH at 25 °C.
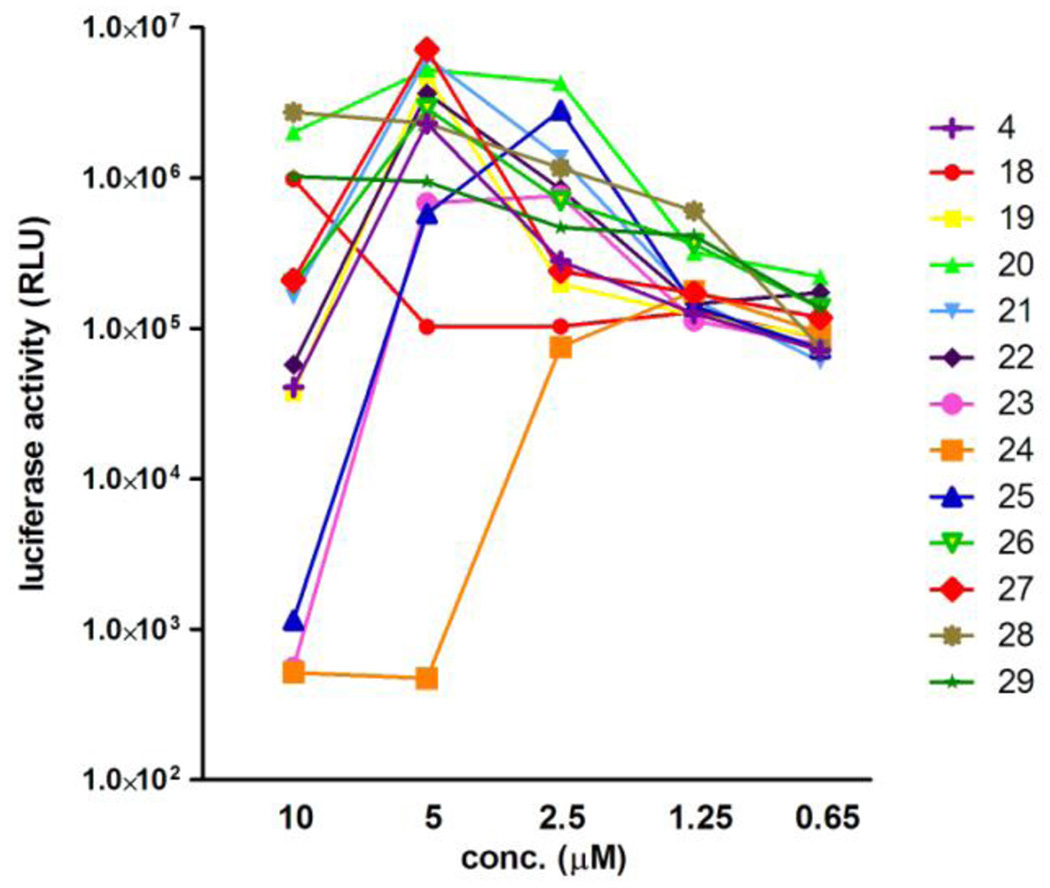
In order to identify the amino acid residues responsible for IN inhibitory and anti-HIV activities of these peptides, an Ala-scan of compound 4 was performed (Figure 6). Compounds 18–22, 25, 27 and 29 showed IN inhibitory activities against the 3’-end processing and strand transfer reactions similar to those of 4 (Table 3). Ala-substitution for Leu7, Gln8, Gln9, Leu10, Leu11, His14, Arg16 or Gly18 did not cause any significant change in either of IN inhibitory activities, indicating that the replaced amino acids are not essential for IN inhibition. Ala-substitution for Phe12, Ile13, Phe15 or Ile17 gave compounds 23, 24, 26 and 28, which were 2–4 times less active in both the IN inhibitory assays, suggesting that Phe12, Ile13, Phe15 and Ile17 are indispensable for IN inhibition. Assessment of anti-HIV activity in the MT-4 Luc system showed that all compounds 18–29 produced dose-dependent inhibition of HIV-1 replication, although they displayed cytotoxicity at 10 µM (4, 19–23, 26 and 27) or above 5 µM (24 and 25) (Figure 7). Compounds 23 and 24, with Ala-substitution for Phe12 and Ile13, respectively, showed weaker inhibitory activity than 4 at 5 µM. Consequently, Phe12 and Ile13 were deemed to be critical for activity, which is consistent with the IN inhibitory activity results. A control peptide isomer of 5 (Ac-QIFEHLAGIIQILRFLRI-R8-NH2) did not show anti-HIV activity at concentrations below 10 µM, suggesting that the original Vpr-sequence, with the exceptions of Phe12, Ile13, Phe15 and Ile17, is critical for activity.
Figure 6.
Amino acid sequences of compounds 18–29 based on an Ala-scan of compound 4. Position numbers correspond to alignment with compound 5.
Table 3.
IC50 values of compounds 18–29 toward the 3’-end processing and strand transfer reactions catalyzed by HIV-1 IN.
| compound | IC50 (µM) | |
|---|---|---|
| 3’-end processing | strand transfer | |
| 4 | 0.11 ± 0.03 | 0.05 ± 0.01 |
| 18 | 0.12 ± 0.004 | 0.08 ± 0.01 |
| 19 | 0.13 ± 0.02 | 0.06 ± 0.01 |
| 20 | 0.10 ± 0.004 | 0.06 ± 0.01 |
| 21 | 0.12 ± 0.02 | 0.07 ± 0.01 |
| 22 | 0.13 ± 0.003 | 0.06 ± 0.01 |
| 23 | 0.34 ± 0.06 | 0.18 ± 0.03 |
| 24 | 0.33 ± 0.02 | 0.22 ± 0.01 |
| 25 | 0.13 ± 0.01 | 0.06 ± 0.01 |
| 26 | 0.25 ± 0.02 | 0.12 ± 0.01 |
| 27 | 0.11 ± 0.01 | 0.05 ± 0.01 |
| 28 | 0.20 ± 0.03 | 0.16 ± 0.02 |
| 29 | 0.09 ± 0.01 | 0.09 ± 0.01 |
Figure 7.
Luciferase signals in MT-4 Luc cells infected with HIV-1 in the presence of various concentrations of compounds 18–29. Luciferase activity was valued as RLU.
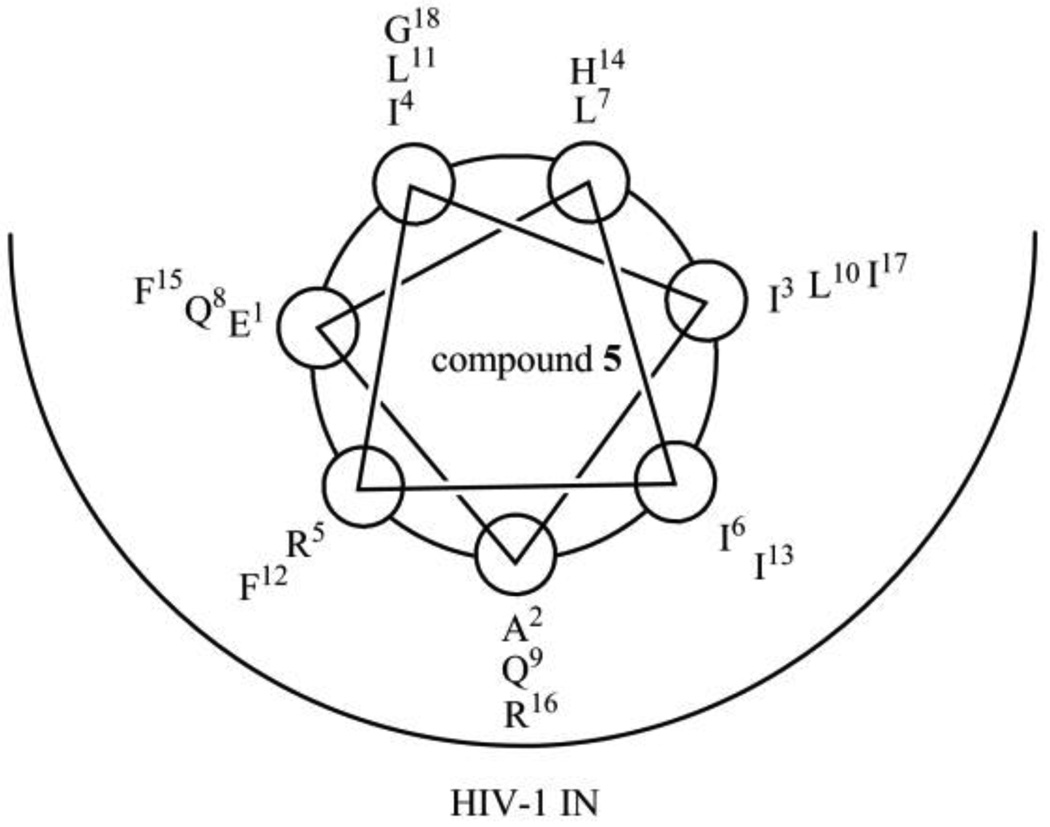
The assumption that compound 5 forms an α-helical structure when binding to HIV-1 IN suggests the binding model of IN and 5 shown in Figure 8, as 5 forms an α-helical structure in 50% aqueous MeOH solution. In this model, Phe12, Ile13, Phe15 and Ile17, which were identified by the Ala-scan experiment as critical residues, are located in the pocket of IN. His14 and Gly18, which can be replaced by Glu-Lys with an increase of activity in compound 11, are located outside of the pocket of IN. Ile3 and Leu7 can also be replaced by Glu-Lys while retaining activity in compound 15, and Leu7 is located outside of the pocket, whereas Ile3 is located in the edge of the pocket. Compounds 11 and 15 might form α-helical structures when binding to IN, although 11 or 15 does not show α-helicity in the CD spectrum. Thus, these compounds might retain IN inhibitory activity. This binding model is compatible with the results of structure-activity relationship studies involving Glu-Lys substitution and Ala-scan. The reason for decreases in IN inhibitory and anti-HIV activity of compounds 12 and 17, which show increases of α-helicity, are possibly due to substitution of Glu-Lys for Ala2 and Ile6, which are located in the pocket of IN. The reason for a decrease in activity of compounds 14 and 16, which show increased α-helicity, might be due to substitution of Lys for Gln8 and Phe15, respectively, which are located in the pocket of IN. The reason for decreases in IN inhibitory and anti-HIV activity of compound 13, which also shows a decrease of α-helicity, are possibly due to substitution of Glu-Lys for Gln9 and Ile13, which are located in the pocket of IN.
Figure 8.
Brief presumed drawing of the binding model of HIV-1 IN and compound 5.
3. Conclusion
In the present study, structure-activity relationship studies were performed on Vpr-derived peptides 4 and 5, which had been previously identified as HIV-1 IN inhibitors.4 The Glu-Lys substitution experiments and Ala-scan data suggest that several amino acid residues of 4 and 5 are indispensable for IN inhibitory and anti-HIV activities, and a binding model of IN and 5 were proposed. Furthermore, two novel compounds 11 and 15, which contained Glu-Lys pairs and showed more potent IN inhibitory activities than compound 5, were found. These data including the binding model should be useful for the development of potent HIV-1 IN inhibitors based on Vpr-peptides.
4. Experimental
4.1 Chemistry
All peptides were synthesized by the Fmoc-based solid-phase method. The synthetic peptides were purified by RP-HPLC and identified by ESI-TOF-MS. Fmoc-protected amino acids and reagents for peptide synthesis were purchased from Novabiochem, Kokusan Chemical Co., Ltd. and Watanabe Chemical Industries, Ltd. Protected peptide resins were constructed on NovaSyn TGR resins (0.26 meq/g, 0.025 and 0.0125 mmol scales for Glu-Lys substitution and Ala-scan peptides, respectively). All peptides were synthesized by stepwise elongation techniques. Each cycle involves (i) deprotection of an Fmoc group with 20% (v/v) piperidine/DMF (10 mL) for 15 min and (ii) coupling with 5.0 equiv. of Fmoc-protected amino acid, 5.0 equiv. of diiopropylcarbodiimide (DIPCI) and 5.0 equiv. of 1-hydroxybenzotriazole monohydrate (HOBt․H2O) in DMF (3 mL) for 90 min. N-Terminal α-amino groups of Glu-Lys substitution and Ala-scan peptides were acetylated with 100 equiv. of acetic anhydride in DMF (10 mL). Cleavage from the resin and side chain deprotection were carried out by stirring for 1.5 h with m-cresol (0.25 mL), thioanisole (0.75 mL), 1,2-ethanedithiol (0.75 mL) and TFA (8.25 mL). After removal of the resins by filtration, the filtrate was concentrated under reduced pressure, the crude peptides were precipitated in cooled diethyl ether and purified by preparative RP-HPLC on a Cosmosil 5C18-AR II column (10 × 250 mm, Nacalai Tesque, Inc.) with a LaChrom Elite HTA system (Hitachi). The HPLC solvents employed were water containing 0.1% TFA (solvent A) and acetonitrile containing 0.1% TFA (solvent B). All peptides were purified using a linear gradient of solvents A and B over 30 min at a flow rate of 3 cm3 min−1. The purified peptides were identified by ESI-TOF-MS (Brucker Daltonics micrOTOF-2focus) (shown in Table S1 in Supplementary data,). All peptides were obtained after lyophilization as fluffy white powders of the TFA salts. The purities of these peptides were checked by analytical HPLC on a Cosmosil 5C18-ARII column (4.6 × 250 mm, Nacalai Tesque, Inc.) eluted with a linear gradient of solvents A and B at a flow rate of 1 cm3 min−1, and eluted products were detected by UV at 220 nm (shown in Figures S1–S3 in Supplementary data).
4.2 Expression and purification of F185K/C280S HIV-1 integrase from E. coli
Plasmid encoding IN1–288/F185K/C280S was expressed in Escherichia coli strain C41. The solubility of the mutant protein was examined in a crude cell lysate, as follows. Cells were grown in 1 L of culture medium containing 100 µg/mL of ampicillin at 37°C until the optical density of the culture at 600 nm was between 0.4 and 0.9. Protein expression was induced by the addition of isopropyl-1-thio-β-D-galactopyranoside to a final concentration of 0.1 mM. After 2 h, the cells were collected by centrifugation at 6,000 rpm for 30 min. After removal of the supernatant, the cells were resuspended in HED buffer (20 mM HEPES, pH 7.5, 1 mM EDTA, 1 mM DTT) with 0.5 mg/mL lysozyme and stored on ice for 30 min. The cells were sonicated until the solution exhibited minimal viscosity then it was centrifuged at 15,000 rpm for 30 min. After removal of the supernatant, the pellet was dissolved in TNM buffer (20 mM Tris/HCl, pH 8.0, 1 M NaCl, 2 mM 2-mercaptoethanol) with 5 mM imidazole and stored on ice for 30 min. The cells were then centrifuged at 15,000 rpm for 30 min and the supernatant was collected. The supernatant was then filtered through 0.45 µm filter cartridge and applied to a HisTrap column at 1 mL/min flow rate. After loading, the column was washed with 10 volume of TNM buffer with 5 mM imidazole. Protein was eluted with a linear gradient of 500 mM imidazole, containing TNM buffer. Fractions containing IN were pooled and checked with SDS-PAGE.
4.3 CD spectroscopy of peptides with Glu-Lys substitution
CD measurements were performed on a JASCO J720 spectropolarimeter equipped with thermo-regulator (JASCO Corp., Ltd.), using 5 µM of peptides dissolved in 0.1 M phosphate buffer, pH 5.6 containing 50% MeOH. UV spectra were recorded at 25 °C in a quartz cell 1.0 mm path length, a time constant of 1 s, and a 100 nm/min scanning speed with 0.1 nm resolution.
4.4 Integrase assays
Expression and purification of the recombinant IN in Escherichia coli were performed as previously reported with addition of 10% glycerol to all buffers. Oigonucleotide substrates were prepared as described.6 Integrase reactions were performed in 10 µL with 400 nM of recombinant IN, 20 nM of 5’-end [32P]-labeled oligonucleotide substrate and inhibitors at various concentrations. Solutions of 10% DMSO without inhibitors were used as controls. Reaction mixtures were incubated at 37 °C (60 min) in buffer containing 50 mM MOPS, pH 7.2, 7.5 mM MgCl2, and 14.3 mM 2-mercaptoethanol. Reactions were stopped by addition of 10 µL of loading dye (10 mM EDTA, 98% deionized formamide, 0.025% xylene cyanol and 0.025% bromophenol blue). Reactions were then subjected to electrophoresis in 20% polyacrylamide–7 M urea gels. Gels were dried and reaction products were visualized and quantitated with a Typhoon 8600 (GE Healthcare, Little Chalfont, Buckinghamshire, UK). Densitometric analyses were performed using ImageQuant from Molecular Dynamics Inc. The concentrations at which enzyme activity was reduced by 50% (IC50) were determined using “Prism” software (GraphPad Software, San Diego, CA) for nonlinear regression to fit dose-response data to logistic curve models.
4.5 Replication assays (MT-4 luciferase assays)
MT-4 luciferase cells (1 × 103 cells) grown in 96-well plates were infected with HIV-1HXB2 in the presence of various concentrations of peptides. At 6–7 days post-infection, cells were lysed and the luciferase activities were measured using the Steady-Glo assay kit (Promega), according to the manufacturer’s protocol. Chemiluminescence was detected with a Veritas luminometer (Promega).
Supplementary Material
Acknowledgments
N.O. and T.T. are supported by JSPS research fellowships for young scientists. This work was supported by Mitsui Life Social Welfare Foundation, Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Health and Labour Sciences Research Grants from Japanese Ministry of Health, Labor, and Welfare, and by the NIH Intramural Program, Center for Cancer Research, US National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi: .bmc. .
References and notes
- 1.Mitsuya H, Erickson J. In: Textbook of AIDS Medicine. Merigan TC, Bartlett JG, Bolognesi D, editors. Baltimore: Williams & Wilkins; 1999. pp. 751–780. [Google Scholar]
- 2.(a) Cahn P, Sued O. Lancet. 2007;369:1235. doi: 10.1016/S0140-6736(07)60571-6. [DOI] [PubMed] [Google Scholar]; (b) Grinsztejn B, Nguyen B-Y, Katlama C, Gatell JM, Lazzarin A, Vittecoq D, Gonzalez CJ, Chen J, Harvey CM, Isaacs RD. Lancet. 2007;369:1261. doi: 10.1016/S0140-6736(07)60597-2. [DOI] [PubMed] [Google Scholar]
- 3.(a) Farnet CM, Bushman FD. Cell. 1997;88:483. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]; (b) Chen H, Engelman A. Proc. Natl. Acad. Sci. USA. 1998;95:15270. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Gleenberg IO, Herschhorn A, Hizi A. J. Mol. Biol. 2007;369:1230. doi: 10.1016/j.jmb.2007.03.073. [DOI] [PubMed] [Google Scholar]; (d) Gleenberg IO, Avidan O, Goldgur Y, Herschhorn A, Hizi A. J. Biol. Chem. 2005;280:21987. doi: 10.1074/jbc.M414679200. [DOI] [PubMed] [Google Scholar]; (e) Hehl EA, Joshi P, Kalpana GV, Prasad VR. J. Virol. 2004;78:5056. doi: 10.1128/JVI.78.10.5056-5067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Tasara T, Maga G, Hottiger MO, Hubscher U. FEBS Lett. 2001;507:39. doi: 10.1016/s0014-5793(01)02945-3. [DOI] [PubMed] [Google Scholar]; (g) Gleenberg IO, Herschhorn A, Goldgur Y, Hizi A. Arch. Biochem. Biophys. 2007;458:202. doi: 10.1016/j.abb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki S, Urano E, Hashimoto C, Tsutsumi H, Nakahara T, Tanaka T, Nakanishi Y, Maddali K, Han Y, Hamatake M, Miyauchi K, Pommier Y, Beutler JA, Sugiura W, Fuji H, Hoshino T, Itotani K, Nomura W, Narumi T, Yamamoto N, Komano JA, Tamamura H. J. Med. Chem. 2010;53:5356. doi: 10.1021/jm1003528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki T, Futaki S, Niwa M, Tanaka S, Ueda K, Sugiura Y. J. Biol. Chem. 2002;277:2437. doi: 10.1074/jbc.M110017200. [DOI] [PubMed] [Google Scholar]
- 6.(a) Yan H, Mizutani TC, Nomura N, Tanaka T, Kitamura Y, Miura H, Nishizawa M, Tatsumi M, Yamamoto N, Sugiura W. Antivir. Chem. Chemother. 2005;16:363. doi: 10.1177/095632020501600603. [DOI] [PubMed] [Google Scholar]; (b) Marchand C, Zhang X, Pais GCG, Cowansage K, Neamati N, Burke TR, Jr, Pommier Y. J. Biol. Chem. 2002;277:12596. doi: 10.1074/jbc.M110758200. [DOI] [PubMed] [Google Scholar]; (c) Semenova EA, Johnson AA, Marchand C, Davis DA, Tarchoan R, Pommier Y. Mol. Pharmacol. 2006;69:1454. doi: 10.1124/mol.105.020321. [DOI] [PubMed] [Google Scholar]; (d) Leh H, Brodin P, Bischerour J, Deprez E, Tauc P, Brochon JC, LeCam E, Coulaud D, Auclair C, Mouscadet JF. Biochemistry. 2000;39:9285. doi: 10.1021/bi000398b. [DOI] [PubMed] [Google Scholar]; (e) Marchand C, Neamati N, Pommier Y. In vitro human immunodeficiency virus type 1 integrase assays. In: Chaires JB, Waring MJ, editors. Methods in Enzymology (Drug-Nucleic Acid Interactions) Vol. 340. Amsterdam: Elsevier Inc.; 2001. pp. 624–633. [DOI] [PubMed] [Google Scholar]
- 7.Morellet N, Bouaziz S, Petitjean P, Roques BP. J. Mol. Biol. 2003;327:215. doi: 10.1016/s0022-2836(03)00060-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.