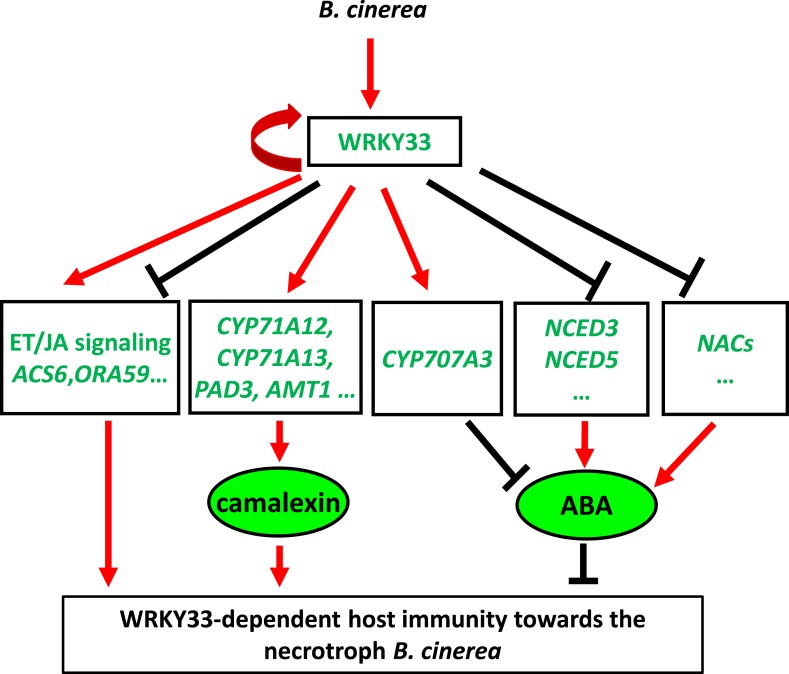
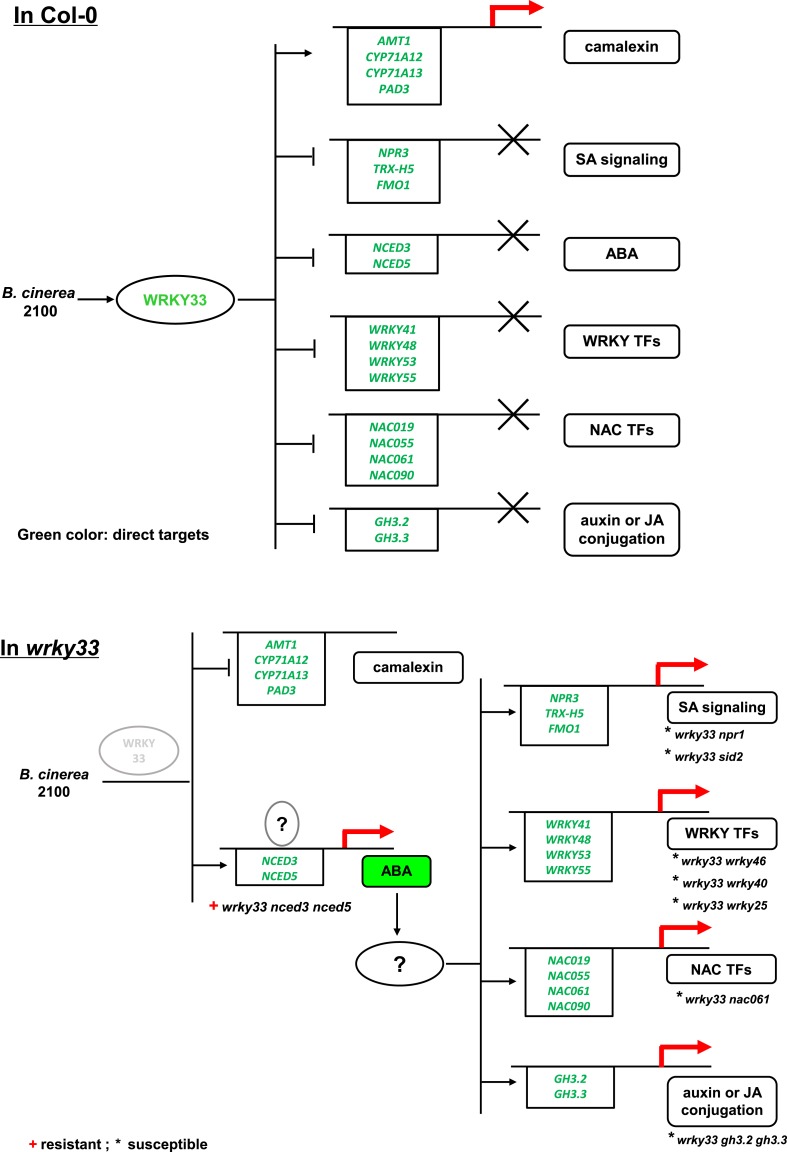
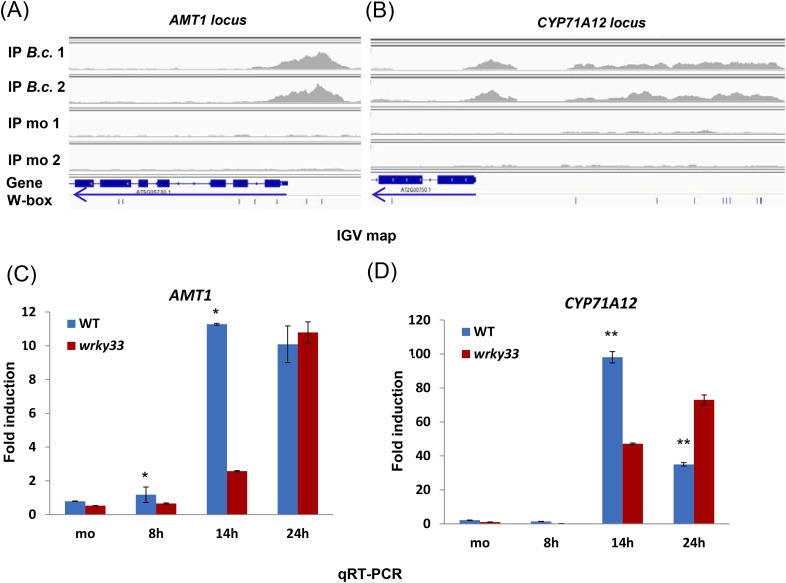
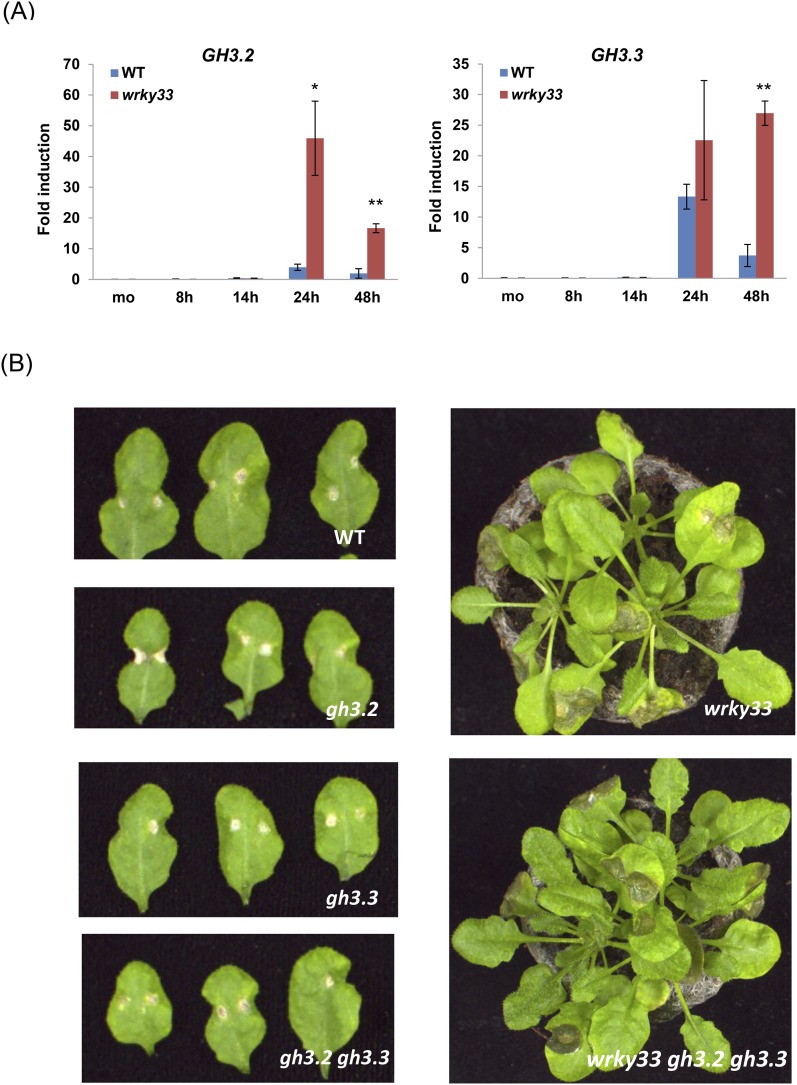
In WT Arabidopsis Col-0 plants (upper panel), B. cinerea inoculation induces WRKY33 expression and protein accumulation that controls ABA levels by repressing the ABA biosynthetic target genes NCED3 and NCED5, and by inducing the ABA metabolic gene CYP707A3. WRKY33 also directly or indirectly represses expression of other genes in different hormone signaling pathways as well as genes encoding WRKY, and NAC TFs. WRKY33 also positively affects resistance towards B. cinerea 2100 by directly targeting and inducing the expression of several genes involved in camalexin biosynthesis. In the wrky33 mutant (lower panel), B. cinerea infection fails to activate host camalexin biosynthesis genes and increases ABA levels by up-regulation of NCED3 and NCED5, which activates downstream ABA-dependent genes and ABA signaling. Among the ABA-dependent genes are genes involved in SA signaling, WRKY TFs, NAC TFs, and genes associated with auxin or JA conjugation indicating that ABA may act as a key sub-node in WRKY33-dependent host immunity to B. cinerea. Whereas WT-like resistance is restored in the wrky33 nced3 nced5 triple mutant, all currently tested mutants of WRKY33-regulated genes acting downstream of ABA failed to restore resistance in the wrky33 mutant background (marked by an asterisk). Thus, additional crucial genes causal for WRKY33-dependent resistance to B. cinerea 2100 remain to be discovered, as do ABA signaling components affecting WRKY33 expression (indicated by the question marks). The solid lines with arrows indicate induction or positive modulation, the bar heads indicate suppression. Direct WRKY33 targets are indicated by the green color.