Figure 6.
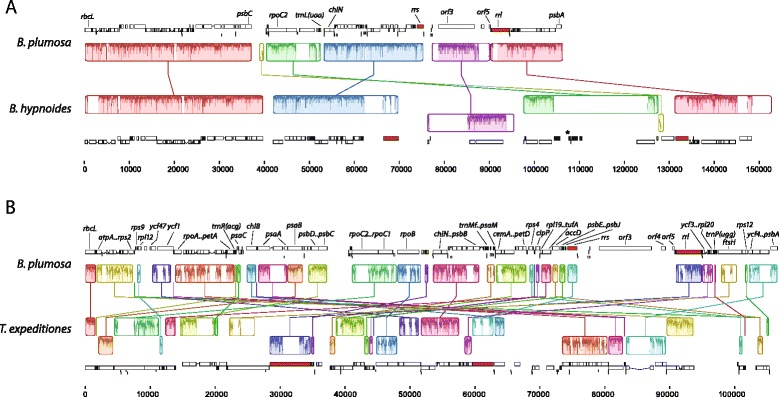
Alignments between the chloroplast genomes of Bryopsis plumosa , B. hypnoides , and Tydemania expeditiones . The Mauve algorithm [81] was used to align the cpDNAs between B. plumosa and B. hypnoides (A), and between B. plumosa and T. expeditiones (B). Corresponding colored boxes are locally collinear blocks (LCBs), which represent homologous regions of sequences that do not contain any major rearrangements. Inside each block a sequence identity similarity profile is shown. Inverted LCBs are presented as blocks below the center line. Annotations are shown above and below the LCBs: protein coding genes as white boxes, tRNA genes in green, rRNA genes in red and short repeats in pink. Lowered position of a box indicates inverted orientation. The asterisk indicates the large tRNA region in B. hypnoides.
