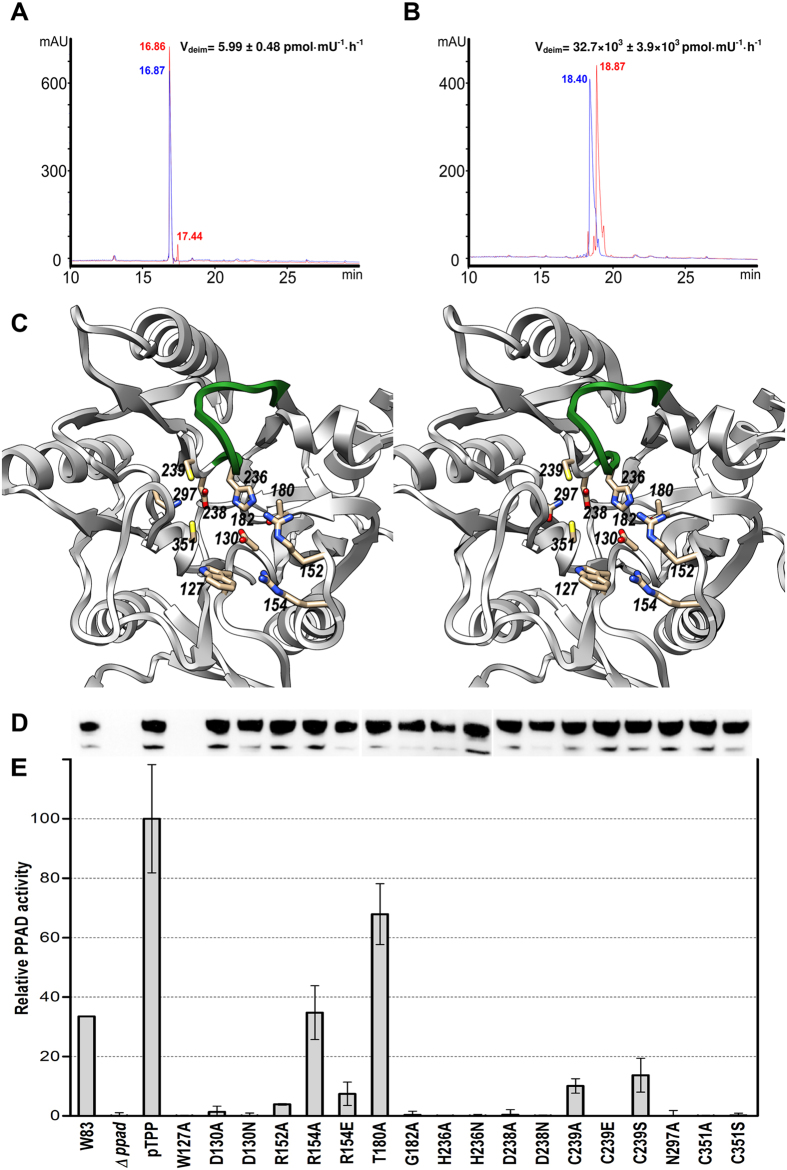
Figure 3. PPAD activity assays.
(A) Endo- and (B) exo-deimininase activity assays in vitro of P. gingivalis W83 wt PPAD against peptides of sequence G-F-S-P-F-R-S-S and P-P-G-F-S-P-F-R, respectively. Peptides are shown before (blue HPLC chromatograms) and after reaction with PPAD (red HPLC chromatograms). Citrullination caused a shift in the retention time of the peptides when compared with the original ones and was confirmed by mass spectrometry. Based on peak integration, the velocity of reaction was calculated for both peptides, which indicated that peptidylarginine exodeiminase activity of PPAD was nearly 5,500 times higher than endodeiminase activity based on reaction velocity (32,700 vs. 6 pmol·mU−1·h−1). (C) Stereo image depicting the 11 positions subjected to point mutagenesis and activity measurements (see (C) and (D)). The Michaelis-loop is shown in green for reference. (D) PPAD expression monitoring through Western-blot analysis of whole bacterial cultures resolved on SDS-PAGE and probed with an anti-PPAD antibody. The samples correspond to those of the abscissa of panel (E). (E) Relative deiminase activity in front of N-acetylarginine of wt W83 strain supernatant (W83), of a PPAD-deletion mutant strain (Δppad), of the latter containing plasmid pTPP for wt PPAD overexpression (pTPP; reference 100%), and a cohort of single point mutants around the active site encoded by pTPP variants.