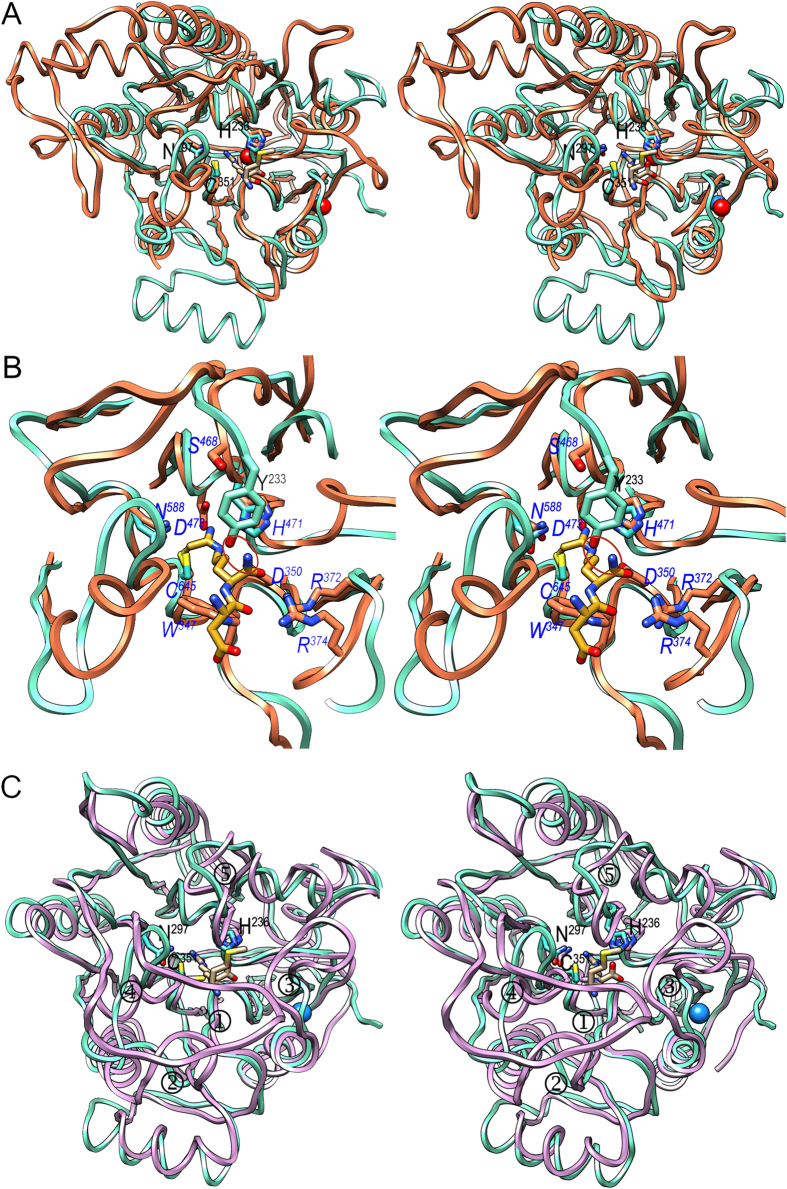
Figure 5. Structural similarities.
(A) Superposed ribbon-plots in stereo of PPAD in its substrate-mimic complex (cyan) and human PAD4 (coral; PDB >4DKT54) as found in its covalent thiouronium reaction intermediate mimic complex. The side chains of the respective catalytic triads (labeled for PPAD only), as well as the two calcium ions of PAD4 (red spheres) and the sodium ion of PPAD (blue sphere) are shown, as is the methionine-arginine dipeptide from the PPAD substrate complex (carbons in tan). Most loops connecting the blades and the consensus secondary elements within each blade differ in length and conformation. (B) Close-up of (A). The side chains of the catalytic triad (not labeled) and Y233 (labeled in black) of PPAD are depicted (carbons in cyan), as are several representative residues from human PAD4 (carbons in coral; labeled in blue italics) and the covalently bound intermediate (carbons in goldenrod). The mechanistically-relevant equivalent positions (see Fig. 4a,b) in PPAD/human PAD4 (in italics) are C351/C645, H236/H471, N297/N588, D238/D473, D130/D350, W217/W347, Y233/S468, R152/R372, and R154/R374. A red ellipse highlights the clash an endodeiminase substrate would have with PPAD Y233. The latter is equivalent to S468 in human PAD4, which allows for free space for C-terminally elongated substrates. (C) Same as (A) showing PPAD (cyan) and AgDI from Enterococcus faecalis (purple; PDB >2JER26) as found in a covalent adduct with an agmatine-derived amidine reaction intermediate. The respective catalytic triads are depicted and that of PPAD is also labeled. AgDI main-chain segments diverging from PPAD and mainly accounting for a closed active site are pinpointed (① to ⑤). The mechanistically-relevant equivalent positions (see Fig. 4a,b) in PPAD/AgDI (in italics) are C351/C357, H236/H218, N297/N306, D238/D220, D130/D96, and W217/W93.