FIG. 1.
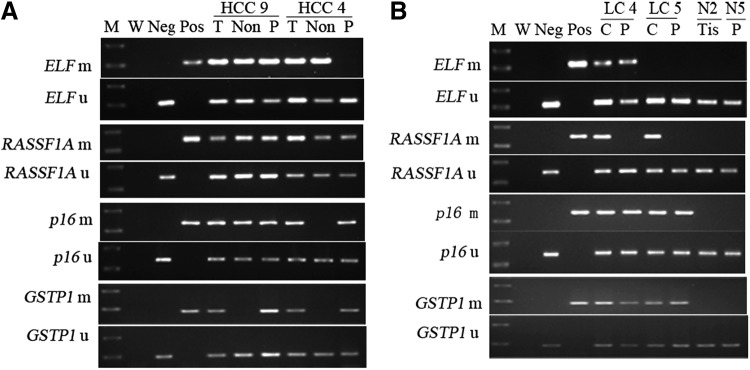
MSP analysis of ELF, RASSF1A, p16, and GSTP1 in liver tissue and plasma samples of hepatocellular carcinoma (HCC) and liver cirrhosis (LC) patients. (A) Representative examples of tumor, nontumor tissue, and corresponding plasma samples from two HCC patients. (B) Methylation status in cirrhosis and corresponding plasma from two LC patients and one normal liver tissue and one plasma sample from patients without liver disease. Bisulfite-treated DNA was amplified with primers specific to the methylated (m) or the unmethylated (u) CpG islands of each gene. MSP products were stained with ethidium bromide after 2.0% agarose gel electrophoresis. C, cirrhosis; M, molecular weight standard; MSP, methylation-specific PCR; m, methylated; W, water contamination control; Pos, MSP product from positive plasma or cell line DNA used as positive control (ELF prom for ELF, Huh-7 for RASSF1A, T47D for p16, and MCF-7 for GSTP1); Neg, unmethylated product of normal liver DNA as negative control; Non, nontumor; N, normal cases; P, plasma samples; PCR, polymerase chain reaction; T, tumor; Tis, tissue; u, unmethylated.
