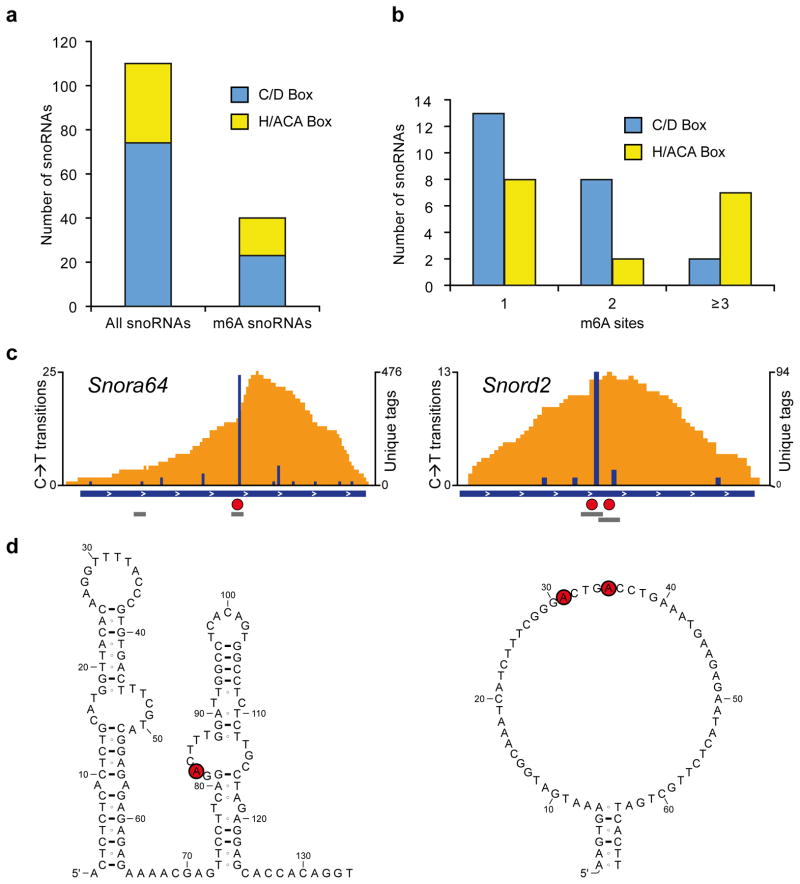
Figure 5. m6As are abundant in mouse snoRNAs.
(a) Bargraph showing the number of snoRNAs analyzed and the number of snoRNAs with at least one m6A. Both, C/D-box and H/ACA-box snoRNAs are methylated.
(b) C/D-box and H/ACA-box snoRNAs can be methylated at multiple positions.
(c) Examples of m6A-modified snoRNAs. The H/ACA-box snoRNA Snora64 and the C/D-box snoRNA Snord2 contain m6A (red circles) in canonical DRACH consensus sites (grey bars). Orange and blue tracks: unique CIMS miCLIP read coverage and C→T transitions, respectively. Horizontal blue bars: transcript models. Grey bars: DRACH consensus sites. Red circles: called m6A residues.
(d) Methylation occurs in single-stranded regions of snoRNAs. Shown are secondary structures of Snora64 and Snord2 as predicted in the Ensembl database. Red circles: called m6A residues.