Abstract
Background
There is an urgent need to improve lung cancer outcome by identifying and validating markers of risk. We previously reported that the cytokinesis-blocked micronucleus assay (CBMN) is a strong predictor of lung cancer risk. Here we validate our findings in an independent external lung cancer population and test discriminatory power improvement of the Spitz risk prediction model upon extension with this biomarker.
Methods
1,506 participants were stratified into a test set of 995 (527 cases /468 controls) from MD Anderson Cancer Center and a validation set of 511 (239 cases / 272 controls) from Massachusetts General Hospital. An epidemiologic questionnaire was administered and genetic instability was assessed using the CBMN assay.
Results
Excellent concordance was observed between the two populations in levels and distribution of CBMN endpoints [binucleated-micronuclei (BN-MN), binucleated-nucleoplasmic bridges (BN-NPB)] with significantly higher mean BN-MN and BN-NPB values among cases (P<0.0001). Extension of the Spitz model led to an overall improvement in the AUC (95% CI) from 0.61 (55.5 – 65.7) with epidemiological variables to 0.92 (89.4 – 94.2) with the addition of BN-MN endpoint. The most dramatic improvement was observed with the never smokers extended model followed by the former and current smokers.
Conclusions
The CBMN assay is a sensitive and specific predictor of lung cancer risk and extension of the Spitz risk prediction model led to an AUC that may prove useful in population screening programs to identify the “true” high risk individuals.
Impact
Identifying high-risk subgroups that would benefit from screening surveillance has immense public health significance.
Keywords: lung cancer, risk model, biomarkers of human exposure
INTRODUCTION
Over 80% of lung cancers are attributed to tobacco exposure; however, fewer than 20% of smokers will develop lung cancer in their lifetime (1– 3). This is a classic example of genetic host susceptibility as a modulator of an exposed individual’s risk for development of cancer (4). Genetically determined modulation of environmental exposures is an attractive mechanism to explain the variation of individual susceptibility among current and former smokers (5). Since biologic variability may occur at any stage of carcinogenesis, the variation in inter-individual susceptibility poses a challenge in quantitative human risk assessment and warrants testing and validation of biomarkers that allow accurate identification of high risk individuals.
A crucial early event in carcinogenesis is the induction of the genomic instability phenotype, which enables an initiated cell to evolve into a cancer cell by achieving a greater proliferative capacity (6). Such instability is mediated through chromosomal changes, at a gross level, and is therefore cytogenetically detected (7). Evidence that cytogenetic biomarkers are positively correlated with cancer risk has been strongly validated in both cohort and nested case-control studies, leading to the conclusion that chromosome aberrations are a relevant marker of cancer risk (8–12), reflecting the outcome of both the genotoxic effects of carcinogens and the genetic host susceptibility.
The cytokinesis-blocked micronucleus assay (CBMN) is a commonly used method for measuring DNA damage (13). This multi-endpoint assay simultaneously assesses DNA damage endpoints in the form of micronuclei, nucleoplasmic bridges and nuclear buds as well as other cellular events such as necrosis, apoptosis and cell proliferation (14). Micronuclei in binucleated cells (BN-MN) are identified as chromosome fragments or whole chromosomes that fail to engage with the mitotic spindle; nucleoplasmic bridges in binucleated cells (BN-NPBs) originate from asymmetrical chromosome rearrangements and/or telomere end-fusions (15, 16) and nuclear buds in binucleated cells (BN-NBUDs) represent a mechanism by which cells remove amplified DNA and are markers of possible gene amplification (17).
We have previously reported in a pilot study and confirmed in a larger study (18–20) that BN-MN and BN-NPB are strong predictors of lung cancer susceptibility with an overall positive predictive value of 96.1 and negative predictive value of 89.7 associated with disease status (19). In the current study, we externally validate our findings in an independent lung cancer population and test the effect of extending an existing lung cancer risk prediction model (21) with the CBMN endpoints. To date, several lung cancer prediction risk models have been developed [Bach, Spitz, Liverpool Lung Project, Prostate Lung Colon Ovary and European Prospective Investigation into Cancer and Nutrition] based on epidemiological risk factors with a wide range of discriminatory power (21–25). Only two of such models have been extended by the addition of biomarkers; however, the resulting improvement in discriminatory power is only modest (26–28). Our data show that the extension of the Spitz model with the BN-MN endpoint leads to a substantial improvement in the discriminatory power of the model among all smoking strata.
MATERIALS and METHODS
Study Participants
A total of 1,506 participants comprised the study test set of 995 study participants (527 cases / 468 controls) and validation set of 511 study participants (239 cases / 272 controls). A complete epidemiologic questionnaire was administered and CBMN assay was conducted. The study was approved by institutional review boards at the University of Texas MD Anderson Cancer Center (MDACC), Kelsey-Seybold Clinics, Massachusetts General Hospital (MGH), and the Harvard School of Public Health. All subjects provided written informed consent for participation.
Lung cancer cases
Lung cancer patients were recruited at both MDACC in Houston and MGH in Boston. A total of 527 non-Hispanic White lung cancer cases from MDACC [recruited between 2005– 2011] were included in the model development and internal validation part of the study. A total of 239 lung cancer patients from MGH [recruited between 2009– 2011] were included as an external validation set. All cases were newly diagnosed, histologically confirmed, lung cancer patients enrolled before initiation of chemotherapy or radiation therapy, with no restriction on age, stage, or histology. Lung cancer patients were treated in the Thoracic Surgery, Thoracic Oncology, or Pulmonary Units at MDACC or MGH. Lung cancer diagnosis was histologically confirmed by a lung pathologist.
Healthy Controls
A total of 740 healthy controls were recruited between 2005–2011 from the Kelsey-Seybold Clinic, a multispecialty physician group located in the Houston metro area. Four-hundred and sixty eight healthy controls were frequency matched to the MDACC lung cancer cases by age (±5 years), sex, ethnicity, and smoking status (never, former, or current) and used for the model development and internal validation part of the study. An independent set of 272 healthy controls, that were mutually exclusive from the internal validation set, were frequency-matched to the MGH lung cancer cases on age (± 5 years), gender and smoking status and used to create a comparison group for the MGH lung cancer cases in the external validation part of the study.
For all participants, former smokers were individuals who had smoked at least 100 cigarettes in their lifetime but quit at least 12 months prior to lung cancer diagnosis (for cases) or prior to the interview (for controls). Current smokers include those currently smoking and “recent quitters” [i.e., those who quit within the last 12 months from diagnosis (for cases) or interview (for controls)]. Data on smoking history include smoking duration, number of cigarettes smoked per day, computed pack-years smoked, and age at smoking initiation for all smokers plus age at smoking cessation and computed years since cessation for former smokers. Smoking duration was determined by subtracting the age at which the participant had started smoking from either the age at which the participant had quit smoking (former smokers) or the participant’s current age (current smokers). Pack-years were calculated by multiplying the smoking duration (in years) by the number of cigarettes smoked per day and then dividing by 20. Time of smoking cessation for former smokers was determined by subtracting the age at which the participant had quit smoking from the participant’s current age. Participants were classified as positive for asbestos or wood dusts (sanding or sawdust) exposures if they had been directly exposed for at least 8 hours/week for a year. Participants were classified as positive for a family history of any cancer if at least two first-degree relatives had cancer and positive for a family history of any smoking-related cancer if at least one first-degree relative had a smoking-related cancer. Participants were also classified by self-reported physician-diagnosed emphysema, COPD or hay fever at any time prior to study entry. The MGH cases did not provide hay fever information; therefore we did not include this variable in our extended model.
Lymphocyte Cultures for CBMN Assay
The CBMN assay was performed following the standard cytokinesis-block technique (29, 30) and duplicate lymphocyte cultures were prepared for each study subject (20–22). Slides were scored blindly following the well-established HUMN Project criteria (29–31). Briefly, a cell was identified as binulceated if it contained two nuclei with intact nuclear membranes, an intact cytoplasmic boundary and an intact cell membrane. An event was recorded as a MN if the morphology was identical to that of the two nuclei but smaller, non- retractile and not connected to the main nucleus. An event was recorded as a nucleoplasmic bridge if the connection between the two nuclei was non-retractile and had the same staining quality as the main nuclei. For each sample, 1000 binucleated cells were evaluated and BN-MN and BN-NPB endpoints were recorded. The assay was conducted blinded to case-control status and run consecutively as cases and controls were enrolled into the study, which should nullify selection bias in assay data availability. All MDACC samples were assayed and scored at MDACC. The MGH samples were assayed at Harvard and the blinded slides were sent to MDACC for scoring.
Statistical Methods
External Validation of the CBMN assay
Statistical analyses were performed using SAS and STATA software. Within each participant group, Fisher’s exact test was used to compare discrete variables such as sex, smoking status, second hand smoke, dust exposure, asbestos exposure, and family history of cancer between the cases and controls. The mean differences of continuous variables (age, number of cigarettes per day, and cessation age) between cases and controls were tested by the Student’s t-test. Wilcoxon rank-sum test was applied to compare the distribution of spontaneous BN-MN and BN-NPB between groups.
Development of Extended Spitz Model
Data from MDACC lung cancer cases and controls were used to develop the CBMN extended models for never, former and current smokers, separately. We retained the original epidemiologic variables of the Spitz model (21) namely: second hand smoke [in never smokers], age quit smoking [in former smokers], pack-years and smoking-related cancers [in current smokers], family history of any cancers [in never & former smokers], emphysema, dust and asbestos exposures and hay fever [in former & current smokers], and fit multiple logistic models with these epidemiologic variables and BN-MN CBMN endpoint, stratified by smoking status. Based on our previous studies showing a high correlation between BN-MN and BN-NPB endpoints, we only used the BN-MN endpoint for model extension (21) and developed separate BN-MN extended risk models for never, former and current smokers.
Validation of the Extended Spitz Model
The MGH lung cancer cases and independent controls were used to validate the extended Spitz model and to evaluate the increase in discriminatory power. For each model, we calculated the area under the receiver operator characteristic curve (AUC). Within each smoking group, we performed pairwise comparisons of the AUCs to evaluate model improvement upon marker addition. To further evaluate improvement in discriminatory power within each smoking stratum, we calculated the increase in improvement in risk prediction using the net reclassification improvement method (32). In addition, we compared the positive predictive value and negative predictive value between the original and extended model, overall and stratified by smoking status.
RESULTS
Subject’s Characteristics
Table 1 summarizes the demographics and matching variables for MDACC and MGH study subjects. Mean age ±SD of MDACC study population was 62 ± 10.8 (cases) and 59 ± 12.1 (controls) (p<0.001). On average, controls were 3 years younger but within the 5-year age matching criterion. Cases were significantly heavier smokers than controls measured by pack years. Self-reported histories of COPD, dust and asbestos exposure were significantly higher among cases. Demographic details for the MGH external validation study subjects and the independent MDACC controls are also summarized in Table 1. Cases (n=239) and controls (n=272) were well matched on age, gender and smoking status. Mean age ±SD was 65 ± 10.7 for cases and 64 ± 11.0 for controls. History of COPD but not dust or asbestos exposure was significantly higher among the cases.
Table 1.
Distribution of matching variables and variables from Spitz risk model among cases and controls in the MDACC and MGH populations
| MDACC Overall (N=995) | MGH Overall (N=511) | |||||
|---|---|---|---|---|---|---|
| Case (N = 527) |
Controls (N =468) |
P | Case (N = 239) |
Controls (N = 272) |
P | |
| Matching Variables | ||||||
| Sex, n (%) Male | 260 (49.3) | 243 (51.9) | 0.951 | 107 (44.8) | 114 (41.9) | 0.515 |
| Age, Mean±SD | 62 ± 10.8 | 59 ± 12.1 | <0.001 | 65 ± 10.7 | 64 ± 11.0 | 0.222 |
| Smoking status, n(%) Never Former Current |
111 (21.1) 221 (41.9) 195 (37.0) |
104 (22.2) 185 (39.5) 179 (38.3) |
0.738 |
37 (15.5) 118 (49.4) 84 (35.1) |
60 (22.1) 122 (44.8) 90 (33.1) |
0.166 |
| Spitz Model Variables | ||||||
| Second Hand Smoke, n(%) among NS |
N=111 86 (77.5) |
N=104 84 (80.8) |
0.553 |
N=37 27 (73.0) |
N=60 47 (78.3) |
0.626 |
| Quitting Age, n(%) among FS <42 42–53 ≥ 54 |
N=221 75 (40.5) 51 (27.6) 59 (31.9) |
N=185 65 (29.4) 68 (30.8) 88 (39.8) |
<0.001 |
N=117 45 (38.5) 30 (25.6) 42 (35.9) |
N=122 63 (51.6) 32 (26.2) 27 (22.1) |
0.045 |
| Emphysema, n(%) among FS & CS |
N=416 91 (21.9) |
N=364 25 (6.8) |
<0.001 |
N=167 31 (18.6) |
N-212 18 (8.5) |
0.004 |
| Dust exposure, n(%) among FS & CS |
N=416 81 (19.5) |
N=364 44 (12.1) |
0.005 |
N=202 26 (12.9) |
N=212 29 (13.7) |
0.809 |
| No Hay Fever, n(%) among FS & CS |
N=416 374 (89.9) |
N=364 312 (85.7) |
0.073 |
NA |
N=212 32 (15.1) |
|
| Asbestos exposure, n(%) among FS & CS |
N=195 59 (30.3) |
N=179 25 (14.0) |
<0.001 |
N=84 14 (16.7) |
N=90 18 (20.0) |
0.571 |
| Family history of cancer n(%) among NS & FS |
N=332 101 (30.4) |
N=289 64 (22.2) |
0.020 |
N=150 51 (34.0) |
N=182 51 (28.0) |
0.240 |
| Family history of smoking related cancers, n(%) among CS |
N=195 75 (38.5) |
N=179 57 (31.8) |
0.181 |
N=80 23 (28.7) |
N=90 23 (25.6) |
0.640 |
| Pack-Years, n(%) among CS <28 28–41.9 42–57.4 ≥ 57.5 |
N=195 36 (18.5) 45 (23.1) 56 (28.7) 58 (29.7) |
N=179 76 (42.5) 38 (21.2) 35 (19.6) 30 (16.8) |
<0.001 |
N=69 18 (26.1) 17 (24.6) 16 (23.2) 18 (26.1) |
N=90 49(54.4) 16 (17.8) 16 (17.8) 9 (10.0) |
0.002 |
P=P value from the Chi-square test of association (for categorical variables) and Student’s test (for continuous variables); SD= standard deviation; NS=Never smokers; FS=Former smokers; CS=Current smokers
Distribution of CBMN-endpoints
MDACC population
Mean CBMN biomarker endpoints, BN-MN and BN-NPB were significantly higher in the cases (Table 2). Overall, the mean ± SD of BN-MN for cases was 3.54 ± 0.99 compared to 1.76 ± 0.83 for controls (p<0.001). Distributions for BN-MN did not vary by age, gender, or smoking status. The mean ± SD of BN-MN among never, former and current smokers cases was 3.56 ± 1.02; 3.54 ± 0.98 and 3.54 ± 0.99 as compared to 1.69± 0.81; 1.73± 0.79 and 1.82 ± 0.87 among controls (P<0.001). Similarly, the overall mean ± SD for BN-NPB among cases was 4.25 ± 0.76 compared to 0.99 ± 0.62 among controls (p<0.001). Distributions for BN-NPB did not vary by age, gender, or smoking status. Mean ± SD of BN-NPB among never, former and current smokers cases was 4.18 ± 0.75; 4.25 ± 0.75 and 4.29 ± 0.77 as compared to 0.94 ± 0.59; 0.97 ± 0.60 and 1.04 ± 0.66 among controls (p<0.001).
Table 2.
Distribution of CBMN endpoints among MDACC and MGH populations.
| MDACC | MGH | |||
|---|---|---|---|---|
| Case (N = 527) |
Controls (N =468) |
Case (N = 239) |
Controls (N = 272) |
|
| BN-MN | Mean±SD | Mean±SD | Mean±SD | Mean±SD |
| Overall | 3.54±0.99 | 1.76±0.83 | 3.60±1.01 | 1.81±0.87 |
|
Age ≤62 >62 |
3.55±0.97 3.54±1.01 |
1.78±0.85 1.73±0.80 |
3.72±1.05 3.52±0.97 |
1.78±0.91 1.82±0.84 |
|
Gender Male Female |
3.52±1.00 3.57±0.98 |
1.71±0.82 1.81±0.83 |
3.64±1.03 3.56±0.99 |
1.73±0.87 1.86±0.87 |
|
Smoking Never Former Current |
3.56 ± 1.02 3.54 ± 0.98 3.54 ± 0.99 |
1.69 ± 0.81 1.73 ± 0.79 1.82 ± 0.87 |
3.62± 1.21 3.61 ± 0.93 3.57 ± 1.02 |
1.70 ± 0.81 1.86 ± 0.86 1.80 ± 0.93 |
| BN-NPB | ||||
| Overall | 4.25±0.76 | 0.99±0.62 | 3.89±1.02 | 1.02±0.66 |
|
Age ≤62 >62 |
4.28±0.73 4.23±0.78 |
0.98±0.57 1.00±0.66 |
3.77±1.15 3.91±0.91 |
1.01±0.63 1.04±0.64 |
|
Gender Males Females |
4.28±0.74 4.22±0.77 |
0.99±0.62 0.99±0.62 |
3.97±0.88 3.83±1.11 |
1.02±0.64 1.03±0.63 |
|
Smoking Never Former Current |
4.18 ± 0.75 4.25 ± 0.75 4.29 ± 0.77 |
0.94 ± 0.59 0.97 ± 0.60 1.04 ± 0.66 |
3.86 ± 0.95 3.94 ± 0.96 3.83 ± 1.12 |
1.03 ± 0.8 1.02 ± 0.62 1.00± 0.61 |
Resulting p-values from comparing endpoints between cases and controls (BN-MN or BN-NPB) overall or stratified by age, gender or smoking status. Mean differences between MDACC cases and controls were significant at the 0.0001 level within all stratum. Mean differences between MGH cases and control were significant at the 0.0001 level within all stratum. Mean BN-MN differences between MDACC cases and MGH cases were not significant (P>0.05) within all stratum. Mean BN-NPB differences between MDACC cases and MGH cases were significant (P<0.05) within all stratum.
MGH population
Similar to the MDACC population, the mean CBMN biomarker endpoints, BN-MN and BN-NPB were significantly higher in cases (Table 2). Overall, mean ± SD of BN-MN for cases was 3.60 ± 1.01; 1.81± 0.87 for controls (p<0.001). Distributions for BN-MN did not vary by age, gender, or smoking status. Mean ± SD of BN-MN among never, former and current smokers was 3.62 ± 1.21; 3.61 ± 0.93 and 3.57 ± 1.02 as compared to 1.70 ± 0.81; 1.86 ± 0.86 and 1.80 ± 0.93 among controls (P<0.001). Mean ± SD BN-NPB endpoint, among cases was 3.89 ± 1.02 compared to 1.02 ± 0.66 among controls (p<0.001). Distributions for BN-NPB did not vary by age, gender, or smoking status. Mean ± SD of BN-NPB among NS, FS and CS cases was 3.86 ± 0.95; 3.94 ± 0.96 and 3.83 ± 1.12 as compared to 1.03 ± 0.80; 1.02 ± 0.62 and 1.00 ± 0.61 among controls (p<0.001).
There was excellent concordance of the BN-MN data between the model building and validation populations. Overall, mean ± SD of BN-MN among cases was 3.60 ± 1.01 as compared to 3.54 ± 0.99 (P=0.498) for the test set (Table 2). MDACC cases had slightly higher values of BN-NPB compared to MGH cases (P>0.05). In both model building and model validation sets, cases had significantly higher (P<0.0001) mean BN-MN and BN-NPB values compared to controls, overall and within each smoking strata.
Multivariable Risk Model [The Extended Spitz Model]
Table 3 shows the results of the extended Spitz risk models [never, former and current smokers] using the model building population. This model was developed to generate estimates of predicted 1-year absolute risk of lung cancer. For the risk factors from the original Spitz model, lung cancer among never smokers was not significantly associated with either exposure to SHS (odds ratio [OR], 1.12) or family history of any cancer (OR, 1.06). For former smokers, none of the original Spitz risk factors were significantly associated with lung cancer such as personal history of emphysema (OR, 2.14), exposure to dust (OR, 1.30), family history of any cancer (OR, 1.2), age of smoking cessation for those who quit between age of 42–53 (OR, 1.23), and those who quit at or over age of 54 (OR = 1.35). For current smokers, the original Spitz lung cancer risk factors associated with lung cancer included personal history of emphysema, and smoking pack years (ORs, 5.77 for heaviest, 3.24 for moderate and 3.66 for lightest smokers respectively). Although, the ORs for dust, Asbestos, and smoking related cancer family history in the new model were on par with that reported in the Spitz Model with OR’s of 1.47, 1.67, and 1.51, these did not achieve statistical significance due to the smaller sample size in our study as compared to that used to derive the Spitz model (23).
Table 3.
Extension of Spitz Lung Cancer Risk Models using CBMN Assay Endpoints among Never, Former and Current Smokers in the Model Building (MDACC) Population.
| Variables | BN-MN model1 |
|---|---|
| OR (95% CI) | |
| Never Smokers | |
| CBMN | 16.72 (9.01–31.02) |
| Second Hand Smoke | 1.12 (0.47–2.68) |
| Family History (≥ 2) 2 | 1.06 (0.47–2.43) |
| Former Smokers | |
| CBMN | 15.78 (10.16–24.51) |
| Emphysema | 2.14 (0.94–4.90) |
| Dusts | 1.30 (0.64–2.64) |
| Family History (≥ 2)2 | 1.25 (0.73–2.13) |
| Quit Age: 42–53 | 1.23 (0.66–2.27) |
| Quit Age: ≥ 54 | 1.35 (0.73–2.47) |
| Current Smokers | |
| CBMN | 11.44 (7.40–17.68) |
| Emphysema | 4.92 (2.16–11.23) |
| Pack Yr.: 28–41.9 | 3.66 (1.72–7.76) |
| Pack Yr.: 42–57.4 | 3.24 (1.52–6.90) |
| Pack Yr.: ≥ 57.5 | 5.77 (2.57–12.96) |
| Dusts | 1.47 (0.69–3.11) |
| Asbestos | 1.67 (0.84,3.32) |
| Family History (≥ 1) 3 | 1.51 (0.84–2.69) |
= BN-MN is modeled as continuous variables
= Individuals with 2 or more 1st degree family members with cancer
= Individuals with 1 or more 1st degree family members with a smoking-related cancer
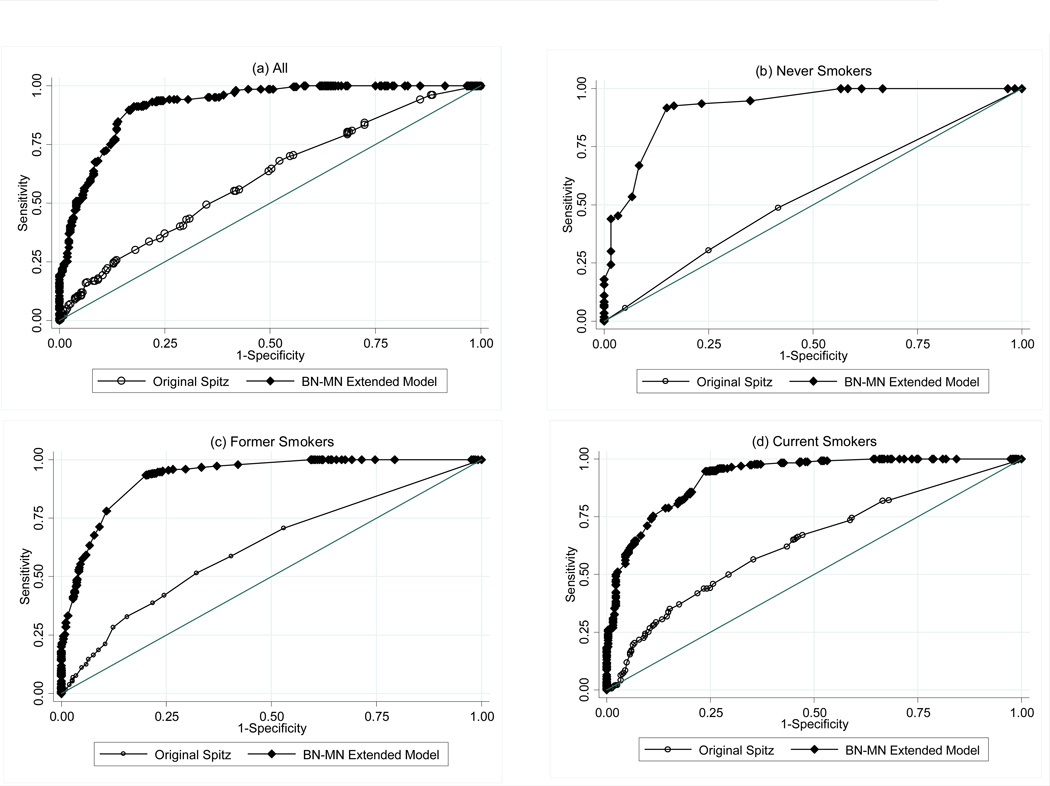
In the extended-model, lung cancer was significantly associated with the BN-MN endpoint as a continuous variable, and the OR’s (95%CI’s) were 16.72 (9.01–31.02), 15.78 (10.16–24.51) and 11.44 (7.40–17.68) for never, former and current smokers respectively. Using the validation set which included the MGH cases we calculated the AUC to estimate the ability of the extended-Spitz model to discriminate between cases and controls. Figure 1 shows the discriminatory power of the extended model overall and by smoking status. Overall, the AUC (95% CI) with the epidemiological variables was 0.61 (55.5 – 65.7), and 0.92 (89.4 – 94.2) with the addition of the BN-MN endpoint. The most dramatic improvement was observed with the never smoker extended model with an increase from 0.55 (44.1 – 66.1) to 0.91 (86.3 – 97.3), followed by the former smoker extended model with an increase from 0.58 (50.5 –65.4) to 0.91(87.3 – 94.8) and the current smoker extended model with an increase from 0.67 (58.2 – 75.1) to 0.93 (88.5 – 96.4). The pairwise comparisons of the receiver operator characteristic curves showed that the extended models for never, former and current smokers were significantly better than the original model (P<0.0001). For the net reclassification improvement analysis, we defined risk categories based on the lower and upper quartiles of predicted risk from our original model based on the 1 year absolute risk proposed by Bach et al. (22): low, intermediate and high with predicted risk <8%, 8%–50% and >50%, respectively, and calculated the increase in improvement in risk prediction of the extended model within each smoking stratum. We observed a significant improvement in classification of 0.540 among cases and 0.354 among controls which resulted in a total significant improvement of 0.894 (P<0.0001) for all participants.
Figure 1. Baseline and CBMN-Extended Model ROC Curves: Overall and by Smoking Status.
- Overall: Spitz Model 60.6% (55.5–65.7); Extended Spitz 91.8% (89.4–94.2)
- Never Smokers: Spitz Model 55.1% (44.1–66.1); Extended Spitz 91.8% (86.3–97.3)
- Former Smokers: Spitz Model 58.0% (50.5–65.4); Extended Spitz 91.0% (87.3–94.8)
-
Current Smoker: Spitz Model 66.7% (58.2–75.1); Extended Spitz 92.5% (88.5–96.4)P<0.0001
Predictive Capability of the Extended Model
Table 4 shows the positive and negative predictive values for the original and BN-MN extended models. For the original model, overall positive and negative predictive values were poor (<60%). With the addition of the BN-MN, the overall positive predictive value increased 35.9 units to 91.1% and the negative predictive value increased 22.8 units to 81.3%. The most notable increase was among never smokers with an increase of 41.7 units in positive predictive value and 26.7 units in negative predictive value.
Table 4.
Positive Predictive and Negative Predictive Value for the original Spitz model versus CMBN extension of the model, overall and stratified by smoking status using the Model Testing (MGH) Population
| Model | PPV(%) | NPV(%) | |
|---|---|---|---|
| Overall | Spitz Model Extended-Spitz with BN-MN |
55.2 91.1 |
58.5 81.3 |
| Never Smokers |
Spitz Model Extended-Spitz with BN-MN |
50.0 91.7 |
58.3 85.0 |
| Former Smokers |
Spitz Model Extended-Spitz with BN-MN |
64.4 94.1 |
45.1 78.7 |
| Current Smokers |
Spitz Model Extended-Spitz with BN-MN |
43.9 86.4 |
76.7 82.2 |
DISCUSSION
In a proof of principle study we reported and confirmed (18–20) that the CBMN assay is a strong predictor of lung cancer risk. Here, we validate the use of the assay as a sensitive marker for lung cancer risk in an independent lung cancer population recruited from a similar study in a different state. The levels of BN-MN observed were similar between the 2 populations, overall and after stratification by age, gender and smoking status. The significantly higher genetic damage observed in cases confirms the role of genetic instability in the carcinogenic process. Our controls were recruited from one state; however, since the measurements between the two case populations were almost identical, there is no reason to expect differences among the control population.
Our group developed the Spitz risk model (21) based on smoking duration, quit-time, occupational exposures, emphysema/COPD, second hand smoke, family cancer history, dust exposure, and hay fever. The validated concordance statistics showed that this model had equivalent to better discriminatory power and more than adequate clinical utility compared to other models (33). Fields et al (34) compared the discriminatory power of 5 existing models and reported that based on epidemiological risk factors alone, the AUCs ranged from 0.57 to 0.84 depending on the study design and smoking status. The Spitz model for former and current smokers was further extended to include markers of DNA repair capacity, mutagen sensitivity (28) and top GWAS identified SNPs (35). The Liverpool Lung Project model was further extended to include the SEZ6L variant (26). However none of these approaches resulted in large gains in discriminatory power; similar to observations noted upon SNP extensions of the Gail model (35, 36).
Several reasons prompted the selection of the CBMN endpoint for extension of the Spitz model. First, chromosome aberrations are well-validated cancer risk biomarkers (37, 8–11). Second, the CBMN is slowly replacing other chromosome aberrations assays since it is a more sensitive multi-endpoint assay. The BN-MN is the most common CBMN endpoint measured by different laboratories worldwide analyzing chromosome aberrations. Third, the CBMN assay is cost effective and is highly appropriate for use in large population screening studies. In addition, Ceppi et al (38) recently reported a good correlation between micronuclei measured by the CBMN assay in lymphocytes and micronuclei measured in oral epithelium which is a target of cigarette-smoke-induced cancer. Our results suggest a substantive increase in the overall predictive ability of the model. The greatest effect was observed among never smokers followed by former and current smokers. Previously extending the Spitz model with host reactivation and mutagen sensitivity assays led to modest increases in the observed AUC (95% CI) from 0.67 (0.63–0.71) to 0.70 (0.66–0.74) in former smokers and from 0.68 (0.64 – 0.72) to 0.73 (0.69 – 0.77) in current smokers (28). Both assays are labor intensive and not suited for wide-scale application. We also observed an improvement in discriminatory power >89% based on the net reclassification improvement criteria as well as improvement in positive and negative predictive values using the extended model. There are several plausible explanations for the superior performance of the CBMN markers. The assay provides data on both genotoxic and cytotoxic cellular events. The CBMN endpoints measure the end-result of parameters that influence genetic instability such as genetic polymorphisms, transcription variation, post-translational modifications, metabolism, genotoxin exposures and malnutrition which also can cause chromosome damage and micronucleus formation. The CBMN assay is a comprehensive measure of events occurring throughout the cell cycle and is unlike other phenotypic assays, such as the mutagen sensitivity, that only measures the DNA damaging effect of a mutagen in the G2 phase and reflects the sensitivity of the cells to exposure to chemicals without consideration of other parameters such as repair. Furthermore, micronuclei are now recognized as mechanisms by which the recently discovered phenomena chromothripsis and chromoanagenesis, may occur. These phenomena are now being recognized as potential major contributors to the initiation and development of human cancer (39, 40). Chromothripsis involves chromosome shattering of one or a few chromosomes leading to a large number of rearrangements while chromoanageneis describes complex genomic rearrangements that occur as a single catastrophic event rather than a series of independent arrangements (41). Both mechanisms involve massive damage to a very small subset of chromosomes or chromosome arms that were found to be confined within micronuclei (40).
Prevention of even 10% of annual deaths from lung cancer would save an estimated 17,000 lives, equivalent to all the annual deaths in the United States from ovarian cancer and almost all the annual deaths from brain cancers. Recent findings from the national lung screening trial showed a 20% reduction in lung cancer mortality among participants screened with CT compared to chest x-rays. However, national lung screening trial results also indicate that to prevent one death from lung cancer, 320 high-risk individuals must be screened (42). The definition of high risk individuals based solely on age and pack years has resulted in unnecessary additional clinical testing and overtreatment. In order to identify the “true high risk” individuals, it is imperative to consider validated biomarkers of risk. Our study showed that the CBMN assay is an exquisitely sensitive and specific predictor of lung cancer risk and that the extension of the Spitz model led to an AUC that may prove useful in population screening programs. Clinically, the ability to identify high-risk subgroups that might benefit from increased screening surveillance that is not appropriate for low-risk individuals, has immense public health significance.
Acknowledgments
This work was supported by the National Cancer Institute grants CA129050 [R. A. El-Zein]; CA092824 [D. Christiani]; CA055769 [M.R. Spitz] and NIEHS Center Grant ES007784.
Footnotes
The authors declare no conflicts of interest, financial or otherwise
References
- 1.American Cancer Society. Cancer Facts and Figures. Atlanta, GA: American Cancer Society, Inc; 2013. [Google Scholar]
- 2.Amos CI, Wu X, Broderick P, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nature Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doll R, Peto R. Cigarette-smoking and bronchial-carcinoma – dose and time relationships among regular smokers and lifelong non-smokers. J Epidemiol Commun Health. 1978;32:303–313. doi: 10.1136/jech.32.4.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann D, Hoffmann I, El Bayoumy K. Less harmful cigarette: a controversial issue. A tribute to Ernst L. Wynder. Chem. Res. Toxicol. 2001;14:767–790. doi: 10.1021/tx000260u. [DOI] [PubMed] [Google Scholar]
- 5.Sellers TA, Potter JD, Bailey-Wilson JE, et al. Lung cancer detection and prevention: evidence for an interaction between smoking and genetic predisposition. Cancer Res. 1992;52:2694–2697. [PubMed] [Google Scholar]
- 6.Fenech M. Chromosomal biomarkers of genomic instability relevant to cancer. Drug Discov Today. 2002;7:1128–1137. doi: 10.1016/s1359-6446(02)02502-3. [DOI] [PubMed] [Google Scholar]
- 7.Solomon E, Borrow J, Goddard AD. Chromosome aberrations and cancer. Science. 1991;254:1153–1160. doi: 10.1126/science.1957167. [DOI] [PubMed] [Google Scholar]
- 8.Liou SH, Lung JC, Chen YH, et al. Increased chromosome-type chromosome aberration frequencies as biomarkers of cancer risk in a blackfoot endemic area. Cancer Res. 1999;59:1481–1484. [PubMed] [Google Scholar]
- 9.Bonassi S, Hagmar L, Stromberg U, et al. Chromosomal aberrations in lymphocytes predict human cancer independently of exposure to carcinogens. European Study Group on Cytogenetic Biomarkers and Health. Cancer Res. 2000;60:1619–1625. [PubMed] [Google Scholar]
- 10.Bonassi S, Znaor A, Norppa H, et al. Chromosomal aberrations and risk of cancer in humans: an epidemiologic perspective. Cytogenet Genome Res. 2004;104:376–382. doi: 10.1159/000077519. [DOI] [PubMed] [Google Scholar]
- 11.Smerhovsky Z, Landa K, Rossner P, et al. Risk of cancer in an occupationally exposed cohort with increased level of chromosomal aberrations. Environ Health Perspect. 2001;109:41–45. doi: 10.1289/ehp.0110941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tucker JD, Preston RJ. Chromosome aberrations, micronuclei, aneuploidy, sister chromatid exchanges, and cancer risk assessment. Mutat Res. 1996;365:147–159. doi: 10.1016/s0165-1110(96)90018-4. [DOI] [PubMed] [Google Scholar]
- 13.Fenech M. The in vitro micronucleus technique. Mutat Res. 2000;455:81–95. doi: 10.1016/s0027-5107(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 14.Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2007;2:1084–1104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 15.Umegaki K, Fenech M. Cytokinesis-block micronucleus assay in WIL2-NS cells: a sensitive system to detect chromosomal damage induced by reactive oxygen species and activated human neutrophils. Mutagenesis. 2000;15:261–269. doi: 10.1093/mutage/15.3.261. [DOI] [PubMed] [Google Scholar]
- 16.Stewenius Y, Gorunova L, Jonson T, et al. Structural and numerical chromosome changes in colon cancer develop through telomere mediated anaphase bridges, not through mitotic multipolarity. Proc Natl Acad Sci USA. 2005;102:5541–5546. doi: 10.1073/pnas.0408454102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenech M. Biomarkers of genetic damage for cancer epidemiology. Toxicology. 2002;181 – 2:411–416. doi: 10.1016/s0300-483x(02)00480-8. [DOI] [PubMed] [Google Scholar]
- 18.El-Zein RA, Schabath MB, Etzel CJ, et al. Cytokinesis-blocked micronucleus assay as a novel biomarker for lung cancer risk. Cancer Res. 2006;66:6449–6456. doi: 10.1158/0008-5472.CAN-06-0326. [DOI] [PubMed] [Google Scholar]
- 19.El-Zein RA, Fenech M, Lopez MS, et al. Cytokinesis-blocked micronucleas cytome assay biomarkers identify lung cancer cases amongst smokers. Cancer Epidemiol Biomakers Prev. 2008;17:1111–1119. doi: 10.1158/1055-9965.EPI-07-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McHugh MK, Lopez MS, Ho CH, et al. Use of the cytokinesis-blocked micronucles assay to detect gender differences and genetic instability in a lung cancer case control study. Cancer Epidemiol Biomakers Prev. 2013;22:135–145. doi: 10.1158/1055-9965.EPI-12-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spitz MR, Hong WK, Amos CI, et al. A risk model for prediction of lung cancer. J Natl Cancer Inst. 2007;99:715–726. doi: 10.1093/jnci/djk153. [DOI] [PubMed] [Google Scholar]
- 22.Bach PB, Kattan MW, Thornquist MD, et al. Variations in lung cancer risk among smokers. J Natl Cancer Inst. 2003;95:470–478. doi: 10.1093/jnci/95.6.470. [DOI] [PubMed] [Google Scholar]
- 23.Cassidy A, Myles JP, van Tongeren M, et al. The LLP risk model: An individual risk prediction model for lung cancer. Br J Cancer. 2008;98:270–276. doi: 10.1038/sj.bjc.6604158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tammemagi CM, Pinsky PF, Caporaso NE, et al. Lung cancer risk prediction: Prostate, lung, colorectal and ovarian cancer screening trial models and validation. J Natl Cancer Inst. 2011;103:1058–1068. doi: 10.1093/jnci/djr173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoggart C, Brennan P, Tjonneland A, et al. A risk model for lung cancer incidence. Cancer Prev Res. 2012;5:834–846. doi: 10.1158/1940-6207.CAPR-11-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raji OY, Agbaje OF, Duffy SW, et al. Incorporation of a genetic factor into an epidemiologic model for prediction of individual risk of lung cancer: the Liverpool Lung Project. Cancer Prev Res. 2010;3:664–669. doi: 10.1158/1940-6207.CAPR-09-0141. [DOI] [PubMed] [Google Scholar]
- 27.Spitz MR, Amos CI, D’Amelio, et al. Re: Discriminatory Accuracy from Single-Nucleotide Polymorphisms in Models to Predict Breast Cancer Risk. J Natl Cancer Inst. 2009;101:1731–1732. doi: 10.1093/jnci/djp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spitz MR, Etzel CJ, Dong Q, et al. An expanded risk prediction model for lung cancer. Cancer Prev Res. 2008;1:250–254. doi: 10.1158/1940-6207.CAPR-08-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fenech M, Morley AA. Measurement of micronuclei in lymphocytes. Mutat Res. 1985;147:29–36. doi: 10.1016/0165-1161(85)90015-9. [DOI] [PubMed] [Google Scholar]
- 30.Fenech M. Cytokinesis-block micronucleus cytome assay. Nature Protocols. 2007;2:1084–1104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 31.Fenech M, Chang WP, Kirsch-Volders M, et al. HUMN Project: detailed description of the scoring criteria for the cytokinesis-block micronucleus assay using isolated human lymphocyte cultures. Mutat Res. 2003;534:65–75. doi: 10.1016/s1383-5718(02)00249-8. [DOI] [PubMed] [Google Scholar]
- 32.Pencina MJ, D'Agostino RB, Sr, D'Agostino RB, et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Statist Med. 2008;27:157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 33.D’Amelio A, Jr, Cassidy A, Asomaning K, et al. Comparison of discriminatory power and accuracy of three lung cancer risk models. Br J Cancer. 2010;103:423–429. doi: 10.1038/sj.bjc.6605759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fields J, Chen Y, Marcus MW, et al. The contribution of risk prediction models to early detection of lung cancer. J Surg Oncol. 2013;108:304–311. doi: 10.1002/jso.23384. [DOI] [PubMed] [Google Scholar]
- 35.Gail MH, Brinton LA, Byar DP, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J Natl Cancer Inst. 1989;81:1879–1886. doi: 10.1093/jnci/81.24.1879. [DOI] [PubMed] [Google Scholar]
- 36.Gail MH. Value of adding single-nucleotide polymorphism genotypes to a breast cancer risk model. J Natl Cancer Inst. 2009;101:959–963. doi: 10.1093/jnci/djp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hagmar L, Bonassi S, Strömberg U, et al. Cancer predictive value of cytogenetic markers used in occupational health surveillance programs. Recent Results Cancer Res. 1998;154:177–184. doi: 10.1007/978-3-642-46870-4_10. [DOI] [PubMed] [Google Scholar]
- 38.Ceppi M, Biasotti B, Fenech M, et al. Human population studies with the exfoliated buccal micronucleus assay: statistical and epidemiological issues. Mutat Res. 2010;705:11–19. doi: 10.1016/j.mrrev.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 39.Holland AJ, Cleveland DW. Chromoanagenesis and cancer: mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med. 2012;18:1630–1638. doi: 10.1038/nm.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crasta K, Ganem NJ, Dagher R, et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482(7383):53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephens PJ, Greenman CD, Fu B, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144(1):27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.NLST: National Lung Screening Trial Research Team. Reduced lung cancer mortality with low-dose computed tomographic screening. N England J Med. 2011:1–15. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]