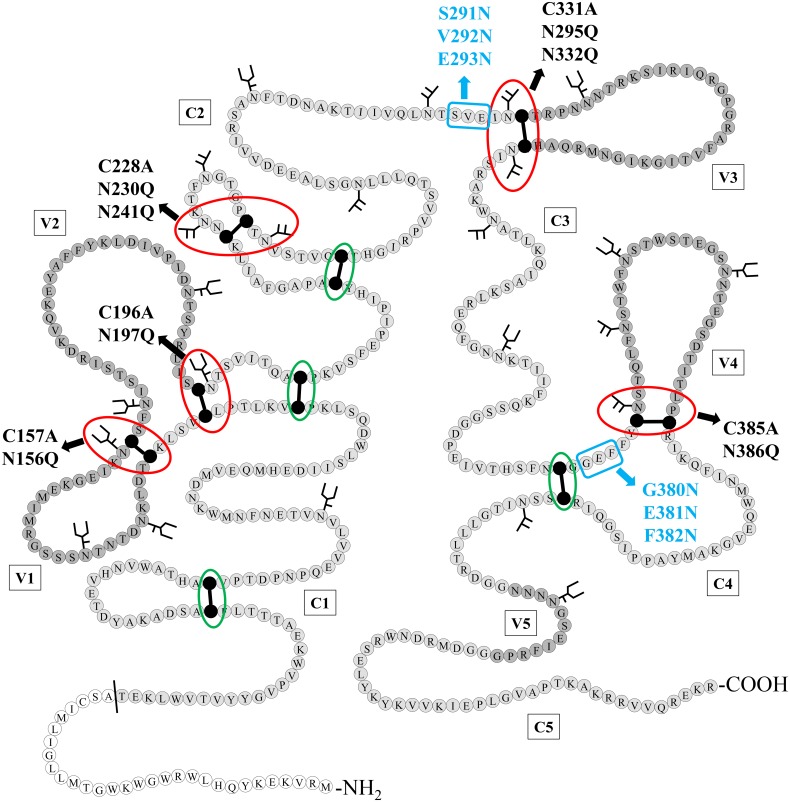
Fig 1. Schematic structure of HIV-1NL4.3 gp120 with indication of the N-glycans and disulphide bridges that were deleted/inserted using site-directed mutagenesis.
Disulphide bridges that were found in close proximity to one or two N-glycosylation sites, either directly neighboring or separated from each other by one other amino acid, are indicated with a red circle. Disulphide bridges that are not directly neighbored by N-glycans are indicated by a green circle. Mutations resulting in deleted disulphide bridges (Cys→Ala) or N-glycosylation sites (Asn→Gln) are indicated in black. Mutations resulting in the generation of novel N-glycosylation sites, due to the introduction of an Asn-X-Ser motif, are indicated in blue (only the glycosylated Asn is shown). The N-terminal signal sequence is colored in white, the conserved domaines (C1–C5) in light grey and the variable loops (V1–V5) in dark grey. Cysteines that are involved in a disulphide bridge are colored black. Figure based on Leonard et al. [3]. Amino acid numbering according to HIV-1 strain HXB2.