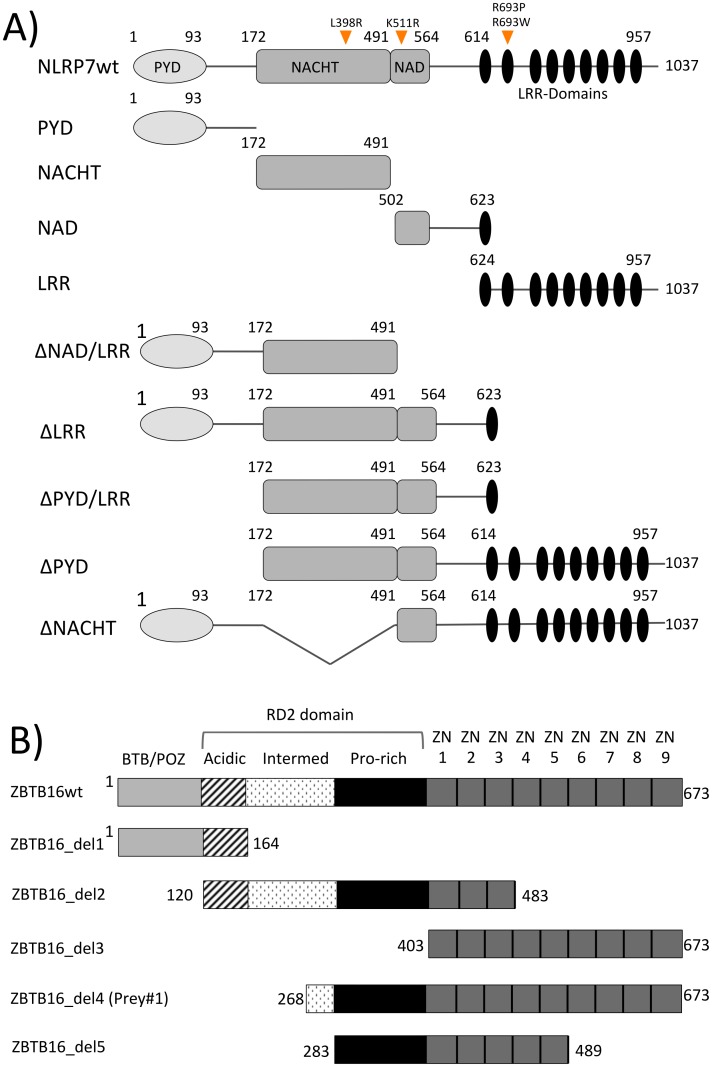
Fig 1. Domain structure of NLRP7 and ZBTB16.
A. Domain structure of NLRP7. Location of analyzed HYDM1-causing mutations L398R, R693P, R693W and non-synonymous variant (NSV) K511R are marked with orange arrowheads. Apart from a full-length construct and the four individual NLRP7 domains, five additional deletion constructs were generated, either lacking one or two domains at a time. B. Domain structure of the transcription factor ZBTB16 identified as interaction partner by the yeast two-hybrid screening. Apart from the full-length construct, containing an N-terminal BTB/POZ, a RD2 and nine zinc-fingers, five additional ZBTB16 deletion constructs were generated (ZBTB16_del1-5). ZBTB16_del4 (aa 268–673) corresponds to “prey#1” identified by the yeast two-hybrid screen.