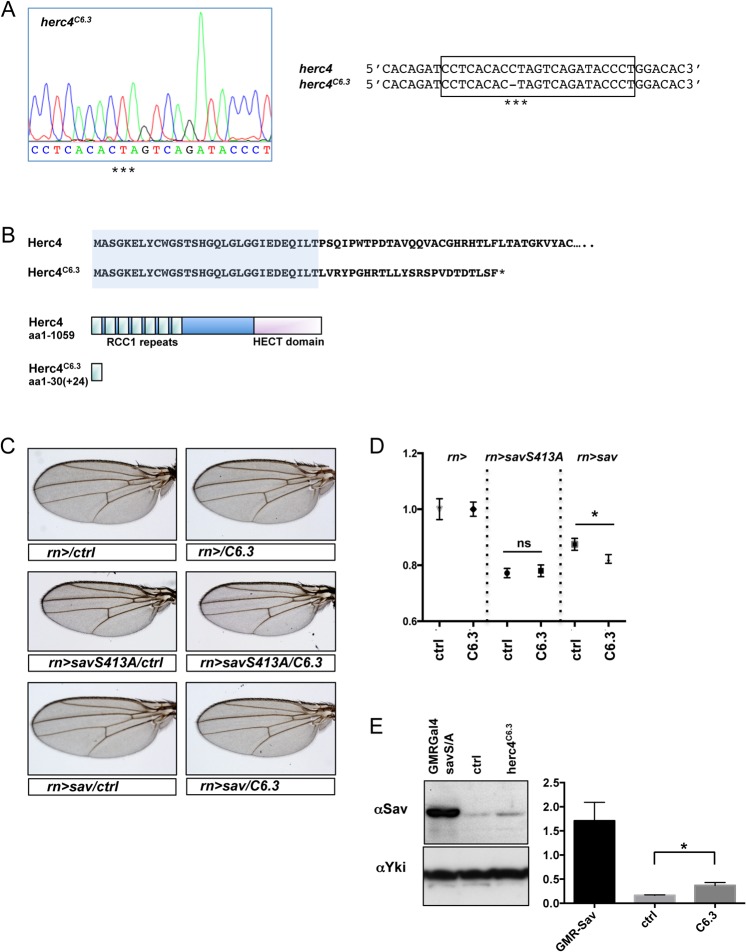
Fig 11. Generation and in vivo analysis of herc4 C6.3 mutant.
(A) Sequence analysis of herc4 CRISPR mutation. Sequence chromatogram of the sgRNA target sequence (boxed in the sequence alignment on the right) showing the loss of cytosine 92 (C92) in the herc4 C6.3 strain (marked by asterisk). (B) Diagram and sequence of the predicted protein resulting from frame-shift mutation. The loss of C92 results in a frame-shift mutation and a truncated protein of 30aa plus 24aa of out-of-frame sequence. (C) herc4 C6.3 enhances the Sav overexpression phenotype in the wing. Both sav (rn>sav) and savS413A (rn>savS413A) transgenes were combined with heterozygosity for either herc4 C6.3 or herc4 ctrl and wings sizes were analysed. (D) Quantification of wing sizes. Shown are fold changes of average wing areas for the indicated genotypes compared to the control (rn/ctrl), which was set as 1. Statistical significance assessed by Student’s t-test. * p < 0.0001. N.S. p > 0.05. n = 10–21. Error bars denote standard deviations. (E) Levels of Sav protein in fly heads of herc4 C6.3, herc4 ctrl and GMR>savS413A were analysed by Western blot. Bottom panel: quantification presented as ratio of Sav/Yki levels from three independent repeats. * p = 0.0385 with a Student’s t-test.