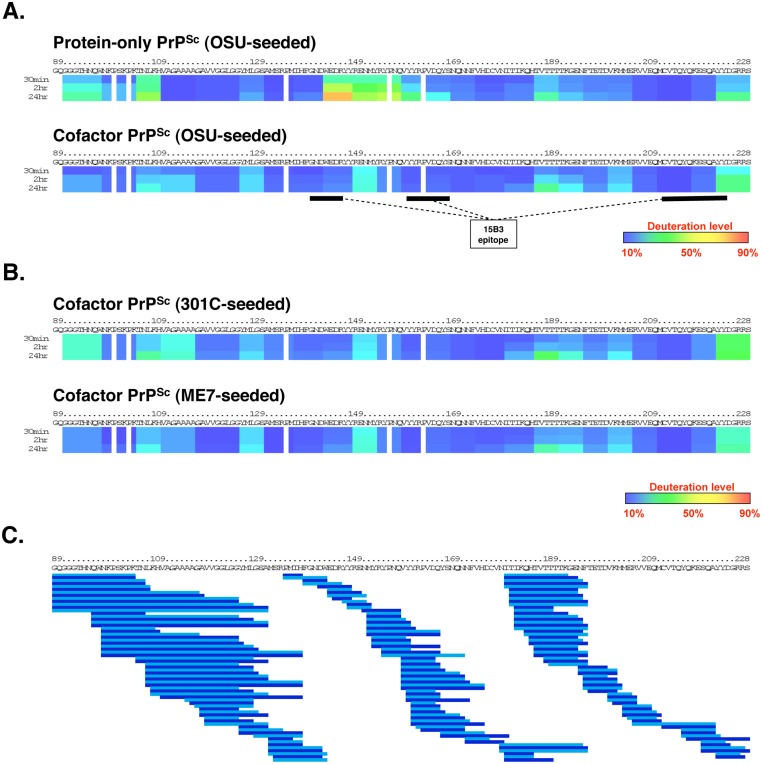
Fig 2. Regional solvent accessibility of cofactor and protein-only PrPSc conformers.
Purified PrPSc conformers were incubated in D2O-containing exchange buffer for 30 min, 2 h, or 24 h and the reaction products were quenched and subjected to LC/MS to measure peptide-specific deuterium incorporation. Data from overlapping peptides was used to quantify localized deuterium exchange and construct ribbon diagrams from a representative experiment. (A) Regional solvent accessibility comparison between cofactor and protein-only PrPSc conformers derived from the same OSU prion strain. The discontinuous epitope of mAb 15B3, which selectively binds infectious PrPSc, is indicated with black bars [18]. (B) Regional solvent accessibility of two additional cofactor PrPSc conformers, derived from the 301C and ME7 prion strains. (C) Map of the 188 high quality deuterated peptides, including different peptide charge states, identified in all four PrPSc experimental samples and used to construct ribbon diagrams in parts (A) and (B). Alternating shades of blue in part (C) are used to highlight neighboring peptides. Despite complete coverage of amino acids 89–230 by overlapping peptides, gaps exist in the ribbon diagrams shown in parts (A) and (B) for two reasons: 1. Deuterium incorporation cannot be quantified for the two N-terminal residues of a peptide as a result of rapid back exchange [19], and 2. Proline residues lack amide hydrogen atoms and therefore do not contribute to deuterium incorporation measurements.