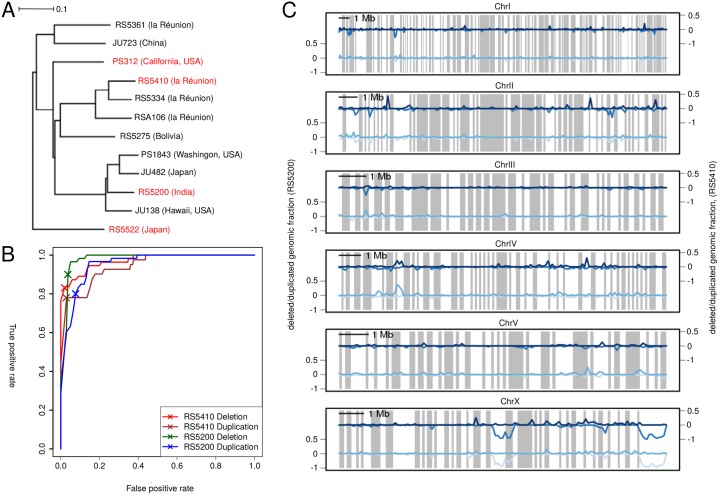
Fig 1. Identification of duplications and deletions in the P. pacificus strains.
A) General phylogeny of P. pacificus strains and the sister species P. exspectatus (RS5522). Strains and species that are used in this study are colored in red. The tree was generated by building a neighbor joining tree on Hamming distances based on roughly 450,000 parsimony informative sites (singletons excluded). Excluding singletons leads to a vast underestimation of distances between strains and species but should increase the robustness of the tree topology. All internal nodes showed a perfect bootstrap support of 100/100. B) True and false positive rate for SV calls for various p-value and fold change cutoffs of the program cnv-seq. The evaluation is based on 100 manually classified SV calls per strain per SV type and final cutoff combinations were chosen subjectively. C) The graphs show the fraction of genomic sequence within non-overlapping 100kb windows, predicted to be duplicated (positive values) and deleted (negative values) relative to the reference strain PS312. Contigs were concatenated using the genetic map of P. pacificus to produce chromosome-scale plots [36]. Gray boxes indicate conserved syntenic regions with the sister species P. exspectatus. While duplications exhibit a much more even distribution, we identified two almost megabase sized regions with high fraction of missing sequence on the X chromosome.