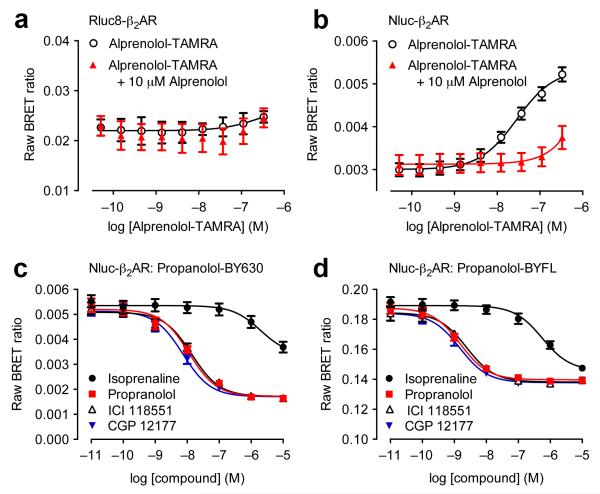
Figure 1.
Suitability of NanoLuc for BRET binding studies. (a,b) BRET ligand binding assays for transiently-transfected Rluc8-β2AR (a) and Nluc-β2AR (b) treated with increasing concentrations of alprenolol-TAMRA in the absence or presence of 10 μM unlabeled alprenolol. Data are mean ± s.e.m. of three experiments performed in quadruplicate. (c,d) Inhibition of the BRET signal for HEK293 cells stably-expressing Nluc-β2AR treated with 10 nM propranolol-BY630 (c) or propranolol-BYFL (d) and increasing concentrations of unlabelled ligands as shown. Each data point represents mean ± s.e.m. of five (all curves in (c) and propranolol in (d)) or four (d) separate experiments. In each experiment we made triplicate determinations for each data point. We calculated KD values indicating the affinity of propranolol-BY630 and propranolol-BYFL (mean ± s.e.m.) from saturation binding assays as 18.9 ± 4.1 nM (n=6) and 42.8 ± 10.8 nM (n=8) respectively. Subsequently, we calculated the respective pKi values indicating the affinity of propranolol, ICI 118551 and CGP12177 from the corresponding IC50 values using the Cheng-Prusoff Equation: 8.13 ± 0.05, 8.04 ± 0.04, 8.32 ± 0.03 (competing with propranolol-BY630) and 8.89 ± 0.09, 8.69 ± 0.14, 8.92 ± 0.03 (competing with propranolol-BYFL). These values (particularly those obtained with propranolol-BYFL) are comparable to those obtained by Baker (2005)20. See text for further discussion of the small differences between values obtained with propranolol-BY630 and propranolol-BYFL.