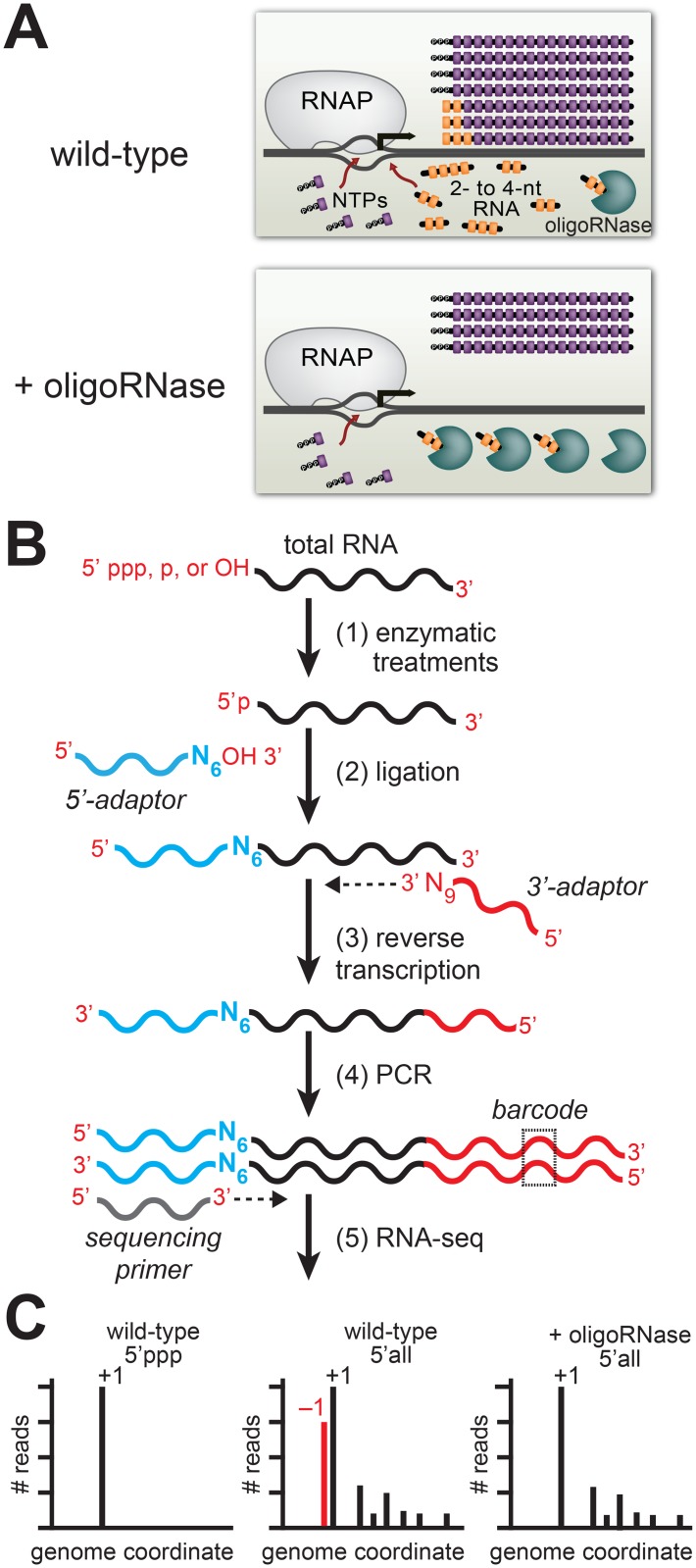
Fig 1. Detection of PDI in bacteria by ectopic expression of an oligoRNase coupled with 5′ RNA-seq.
A. Inhibition of PDI through ectopic expression of an oligoRNase. Depicted is the extent of de novo initiation and PDI from a representative promoter in wild-type cells (top) and cells in which an oligoRNase is ectopically produced (bottom). 2- to 4-nt oligoribonucleotides, are depicted in yellow while NTPs are shown in purple. In the example shown here, PDI leads to generation of full-length transcripts that do not carry a 5′ triphosphate and are one base longer than the products of de novo initiation. B. Steps in 5′ RNA-seq [16]. Step 1: selective enzymatic treatments of total RNA that allow cDNA libraries to be constructed from RNAs on the basis of the phosphorylation state of the 5′ end. Step 2: ligation of 5′ adaptor (in blue) carrying six random bases (N6) at the 3′ end. Step 3: Reverse transcription using a primer with sequence of the Illumina 3′ adaptor on the 5′ end and nine random bases at the 3′ end (N9). Step 4: PCR step includes a primer that introduces a barcode (dashed box) enabling several libraries to be analyzed, in parallel, on the Illumina HiSeq. C. Histograms generated from 5′ RNA-seq. Analysis of transcripts with a 5′ triphosphate (5′ ppp) is used to identify primary start sites (+1). Comparison of results obtained from wild-type cells or cells in which an oligoRNase is ectopically expressed using the analysis of the 5′ ends of all transcripts (i.e. those carrying a 5′-triphosphate, 5′-monophosphate, or 5′-hydroxyl) identifies transcripts generated by PDI that emanate from position −1 (red).