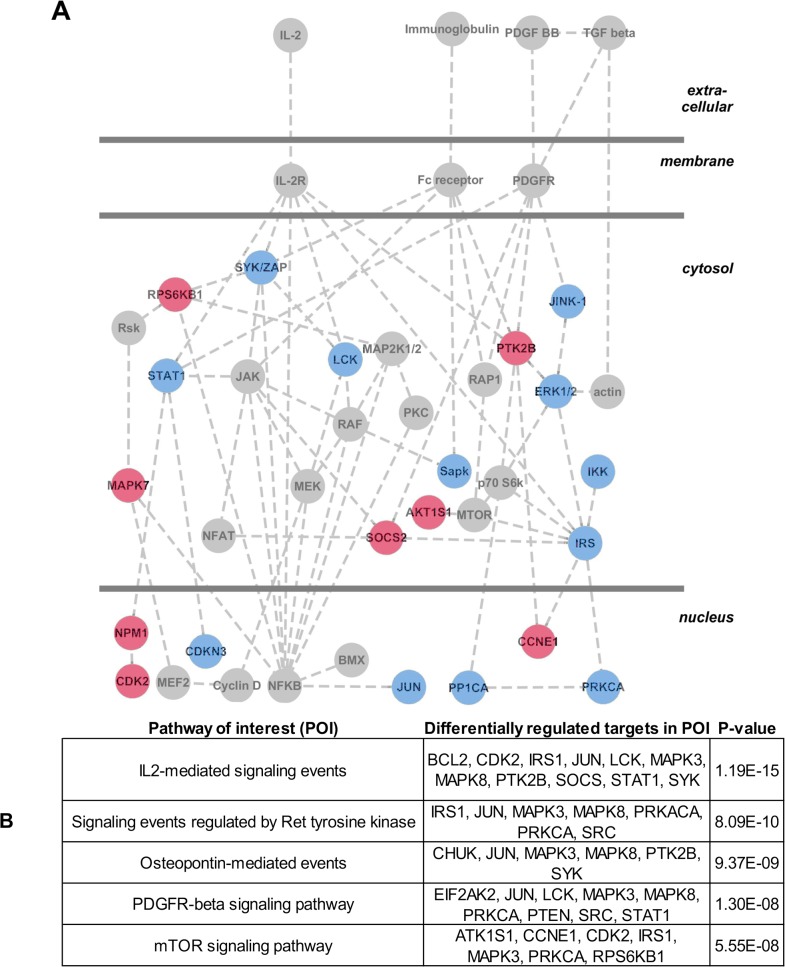
Fig 5. Kinomic screening and prediction of the signalling events that underlie plasmin-mediated immunomodulation.
(A) MoDCs were incubated in the presence/absence of 1 nM t-PA and 100 nM plasminogen. 3 h later cultures were lysed and subject to kinomic microarray analysis yielding a short list of 31 signalling proteins that were differentially regulated upon plasmin-treatment (see S1 Table for complete dataset). The short list was subjected to Ingenuity Pathway Analysis to yield three canonical signalling pathways (not shown) for plasmin-mediated immunomodulation, which were manually merged and refined to generate a single putative signalling pathway that underlies plasmin-mediated immunomodulation. Red symbols represent upregulated events. Blue symbols represent downregulated events. (B) The kinomic short list of differentially regulated signalling proteins was batch analysed using the Pathway Interaction Database (PID). Shown are the 5 most significantly perturbed pathways of interest (POI), the differentially regulated proteins within each POI and the degree of statistical significance (p-value computed using the default PID hypergeometric distribution test).