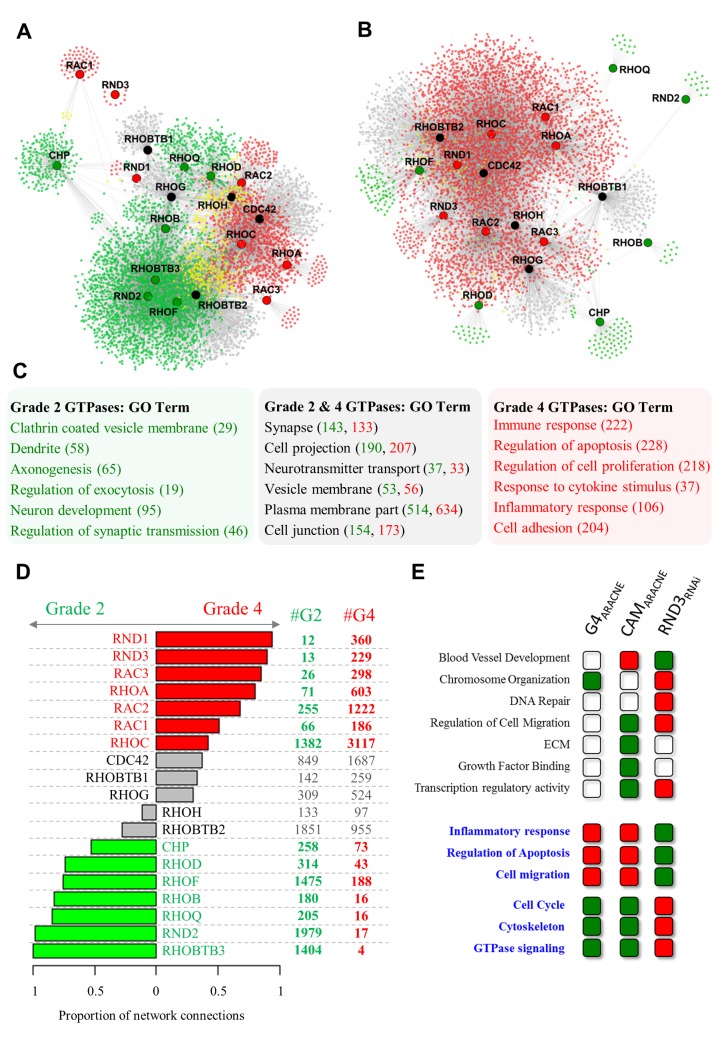
Fig 3. Transcriptional networks reveal two groups of Rho GTPases with divergent connectivity in grade II and grade IV glioma.
A, B. Transcriptional co-expression networks representing the neighbourhood of Rho GTPases in (A) grade II (5973 genes) and (B) grade IV (5621 genes) glioma. C. Gene Ontology terms enriched within genes connected to grade II specific Rho GTPases or grade IV specific Rho GTPases (determined by connectivity, shown in panel D). False discovery rate < 10%, number of genes indicated in brackets (grade II: green, grade IV: red). D. Two distinct subsets of Rho GTPases show a significantly higher number of connections in either grade II (green) or grade IV (red) glioma. Differences in connectivity were expressed as the proportion of grade II to grade IV connections. Rho GTPases with an absolute difference between tumour grades > 0.4 were considered grade specific. E. Transcriptional profiling of U87 siRND3 cells induces a functional profile consistent with the network analysis in primary and CAM implanted tumours. Each Gene Ontology term represents genes within that category which are linked to RND3 (connected to RND3 in the grade IV glioma network and/or connected to RND3 in the CAM ARACNE network and/or differentially expressed in U87 siRND3 cells). These genes are either positively (red boxes) or negatively (green boxes) correlated with expression of RND3 or up-regulated (red boxes) or down-regulated (green boxes) in U87 siRND3 cells. Gene Ontology terms matched to all 3 experiments are highlighted in blue. Genes in networks (A) and (B) are coloured according to whether they are connected to Rho GTPases with grade specific connectivity in grade IV glioma (red nodes), grade II glioma (green nodes), or both grade II and grade IV (yellow nodes). Rho GTPases in networks (A) and (B) are coloured according to the grade specific connectivity shown in panel D (red: grade IV, green: grade II, black: non-specific).