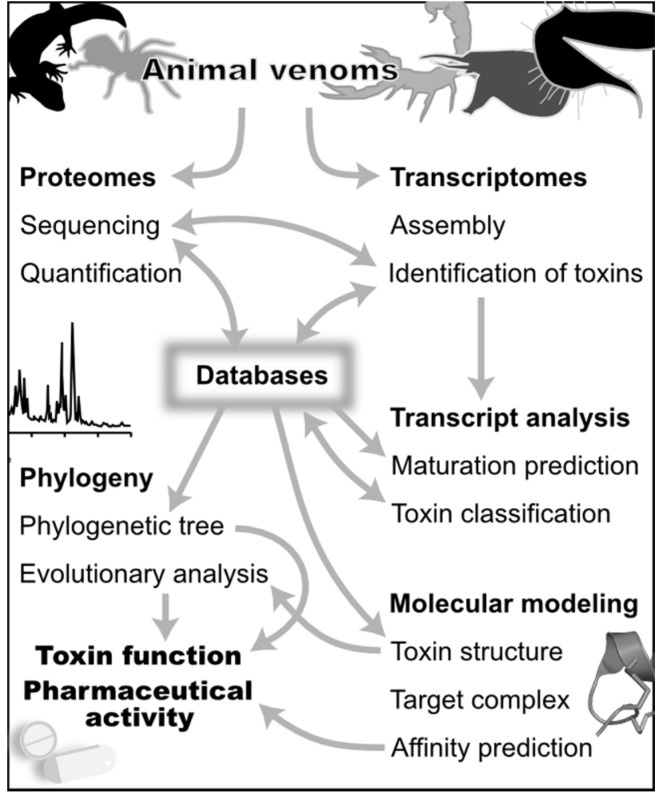
Figure 1.
Computational approaches in venomics. The different themes in this figure are discussed in the text. Generalist and specialized databases are a central source of information for both the discovery (top) and analysis (bottom) of venoms. Venomics computational tools combine data from proteomics and transcriptomics to discover the set of toxins in a venom gland. These tools use predictions of mature toxins from transcriptomes to support peptide sequencing, and sequencing by mass spectrometry uses transcript sequences as a database to rapidly identify peptides and their post-translational modifications. Bioinformatics tools use the standardized classification stored in databases to analyze transcripts, and phylogenetic analyses can be used to analyze toxin evolutionary relationships. Databases in turn record the newly identified toxin peptide and transcript sequences. These sequence data can be also used to refine toxin classification, for example when a new phylogenetic group of toxins is identified. The molecular target of toxins can be suggested by a phylogeny and an evolutionary analysis complemented by the prediction of toxin 3D structures. Molecular modeling of target/toxin complexes can be used to analyze in great depth structure-activity relationships of toxins. Finally, the affinity of these complexes can be predicted from the molecular models.