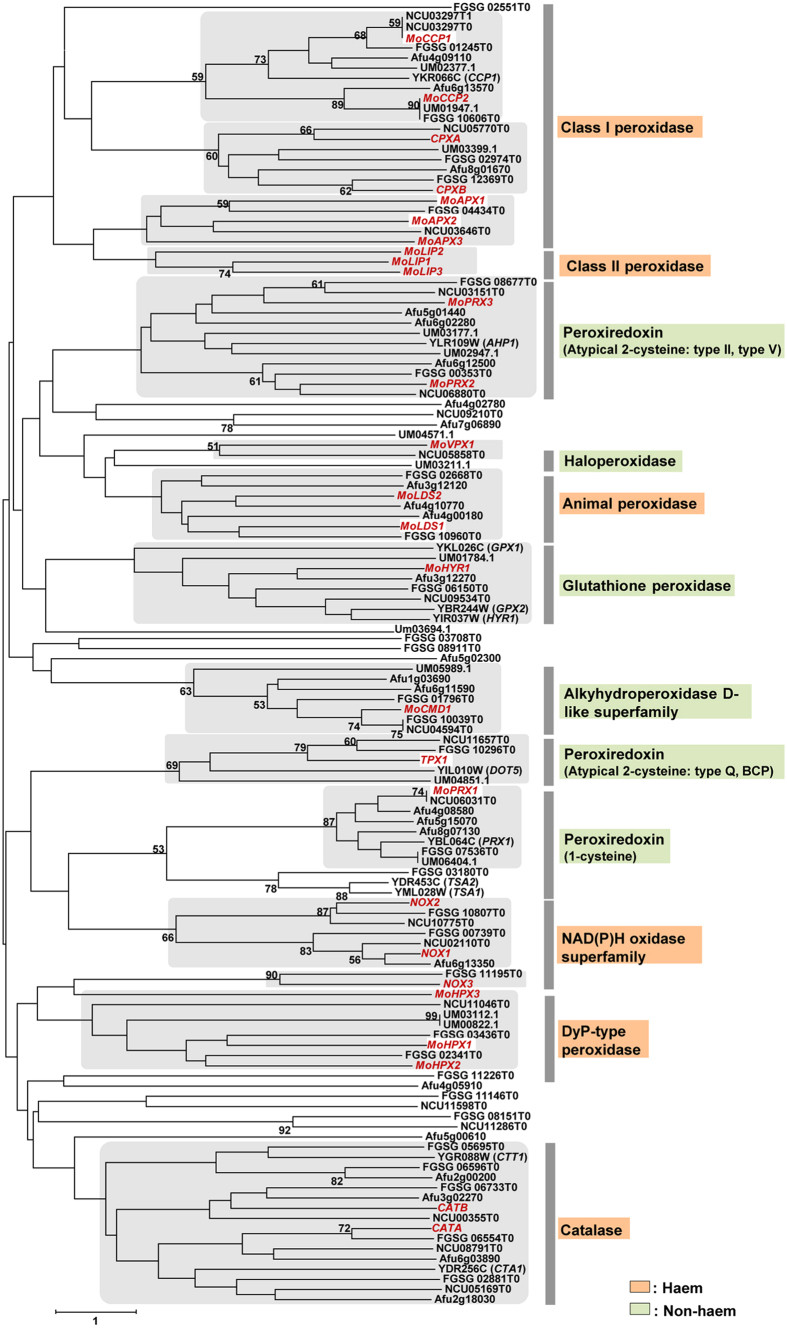
Figure 1. Phylogenetic analysis of putative peroxidase genes in six selected fungi.
A neighbor-joining tree was constructed based on the amino acid sequences of representative fungal peroxidase genes. Numbers at nodes represent bootstrap confidence values, or percentage of clade occurrence in 2,000 bootstrap replicates; only nodes supported by >50% bootstraps are shown. The scale bar represents the number of amino acid differences per site. Sub-clades containing Magnaporthe oryzae peroxidase genes are shaded, in which M. oryzae peroxidase genes and characterized genes from other fungi are depicted in bold red or black, respectively. Abbreviations for fungal species, followed by their GenBank accession numbers, are as follows: Mo, Magnaporthe oryzae; Sc, Saccharomyces cerevisiae; Fg, Fusarium graminearum; Nc, Neurospora crassa; Um, Ustilago maydis; Af, Alternaria fumigatus.