Figure 1.
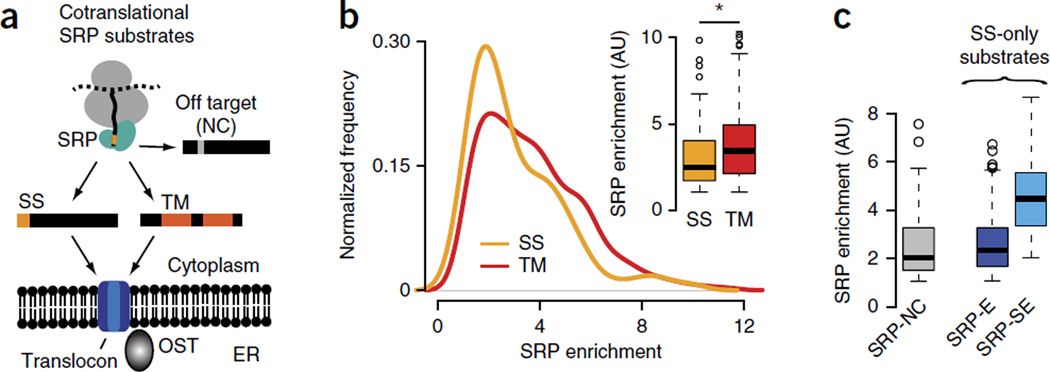
The SRP binds nascent chains in vivo with a broad distribution of specificities. (a) Schematics of selective, cotranslational SRP interaction with proteins bearing signal sequences (SS) or transmembrane (TM) segments. The SRP also binds few noncognate (NC) cytosolic and nuclear off-target proteins that do not get translocated. (b) Enrichment scores of cotranslational SRP binding for SS proteins (i.e., proteins with an N-terminal SS) and for TM proteins (i.e., proteins with TM segments as SRP-binding sites). Inset, significantly higher enrichment of SRP binding in TM proteins (n = 190) compared to SS proteins (n = 107) (P = 0.0007 by Wilcoxon rank-sum test). (c) Classification of SS proteins into noncognate (SRP-NC; n = 109), enriched (SRP-E; n = 78) and strongly enriched (SRP-SE; n = 29) substrates, on the basis of increasing SRP enrichment scores. Box plots show the data distribution through median (center line), first and third quartiles (filled box), 1.5× interquartile range (dashed line) and extreme values (circles). AU, arbitrary units. *P < 0.05.