Figure 5.
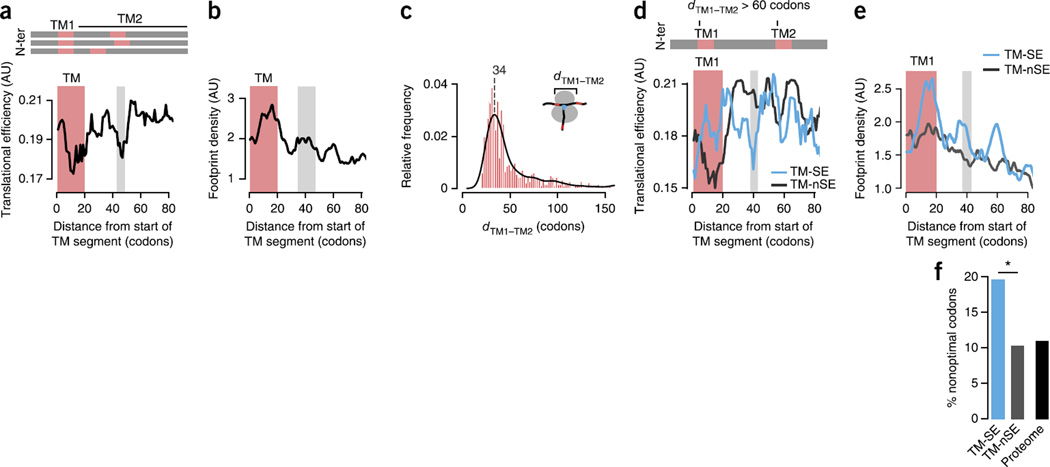
A local slowdown of translation promotes TM-helix recognition by the SRP. (a,b) Median TE (a) and median footprint density (n = 177) (b) profiles for TM proteins (n = 37), showing slower translation of TM segments (red bars) and the region ~40 codons downstream (gray bars). N-ter, N terminus. (c) Distribution of the distance dTM1–TM2 between the start of the first and second TM segments in proteins with multiple TM helices. The most frequent distance is ~34 codons. (d) Median TE profiles for strongly enriched (TM-SE; n = 46) and not strongly enriched (TM-nSE; n = 165) TM proteins with dTM1–TM2 > 60 codons, showing that nonoptimal codons promote SRP recognition independent of the presence of a second TM successive segment. (e) Median ribosome footprint density profiles confirming locally slower translation kinetics for TM-SE (n = 14) substrates compared to TM-nSE (n = 27) substrates. (f) Enrichment in nonoptimal codons at the translational-slowdown element in TM-nSE substrates (P = 0.030 by Fisher’s exact test on counts of optimal and nonoptimal codons; two sided). *P < 0.05.