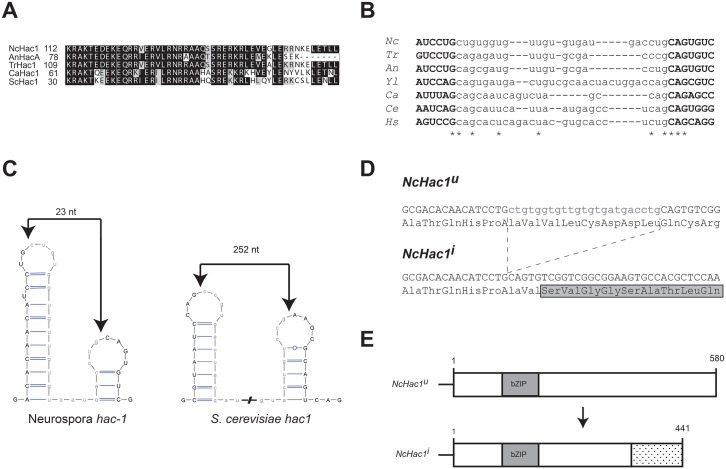
Fig 1. Neurospora crassa has a putative Hac1 homolog.
A) Alignment of the amino acid sequence of the bZIP domain of selected fungal HAC-1 homologs. Nc: Neurospora crassa; An: Aspergillus nidulans; Tr: Trichoderma reseei; Ca: Candida albicans; Sc: Saccharomyces cerevisiae. B) Sequence alignment of the putative non-conventional intron and surrounding regions of hac-1 homologs in fungi. Asterisks denote positions conserved in all the sequences considered in the alignment. Nc: Neurospora crassa; Tr: Trichoderma reseei; An: Aspergillus nidulans; Yl: Yarrowia lipolytica; Ca: Candida albicans; Ce: Caenorhabditis elegans; Hs: Homo sapiens. C) Predicted twin stem-loop structure of the intron of the N. crassa and yeast hac-1 mRNAs. Folding prediction was made with mFOLD [76] and the structures drawn with VARNA [77]. Predicted cleavage sites are indicated by an arrow. The predicted intron sequence is shown in lowercase. D) Nucleotide and deduced amino acid sequence surrounding the putative splice sites in the N. crassa hac-1 CDS. Upon ER stress, the putative intron (shown in lowercase) is removed from the uninduced version of the hac-1 mRNA (hac-1 u), resulting in the induced version (hac-1 i). Translation of the induced version would alter the reading frame, leading to a different C-terminal region. E) Schematic representation of NcHAC-1u and NcHAC-1i.