Figure 3.
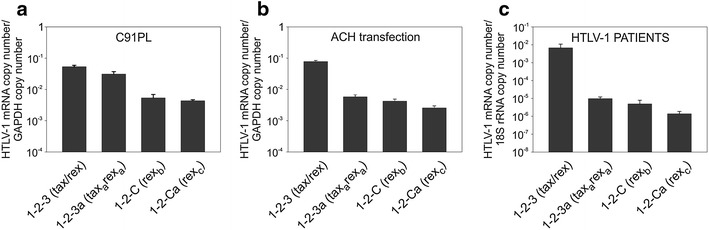
Quantitation of novel alternatively spliced HTLV-1 mRNAs. The tax/rex (1-2-3), tax a /rex a (1-2-3a), rex b (1-2-C) and rex c (1-2-Ca) mRNAs were quantified by qRT-PCR in a the C91PL T-cell line, which is chronically infected with HTLV-1, b HLtat cells 24 h after transfection with the ACH molecular clone, and c Peripheral blood mononuclear cells (PBMC) from three HTLV-1-infected patients (ATLL-2, TSP-3 and TSP-4), after 48 h of culture in vitro. a, b Mean values and standard error bars from three independent experiments. C91PL cells [14] were maintained in RPMI 1640 (Sigma-Aldrich) supplemented with 10% FCS, 2 mM l-glutamine (GIBCO) and penicillin/streptomycin. PBMCs were isolated from peripheral blood samples donated by patients with a clinical diagnosis of ATLL or TSP/HAM attending the clinic at the National Centre for Human Retrovirology, Imperial College Healthcare NHS Trust, St. Mary’s Hospital or King’s College Hospital, London, UK, and treated as previously described [5, 15]. RNA extraction, DNAase treatment, reverse-transcription and qRT-PCR for the detection of HTLV-1 transcripts were performed as previously described [5] by using the primers and probes listed in Table 1. The absolute copy number of each transcript was determined and normalized (normalized copy number, NCN) for the copy number of 18S rRNA (patients’ samples) or GAPDH (C91PL and ACH transfections).
