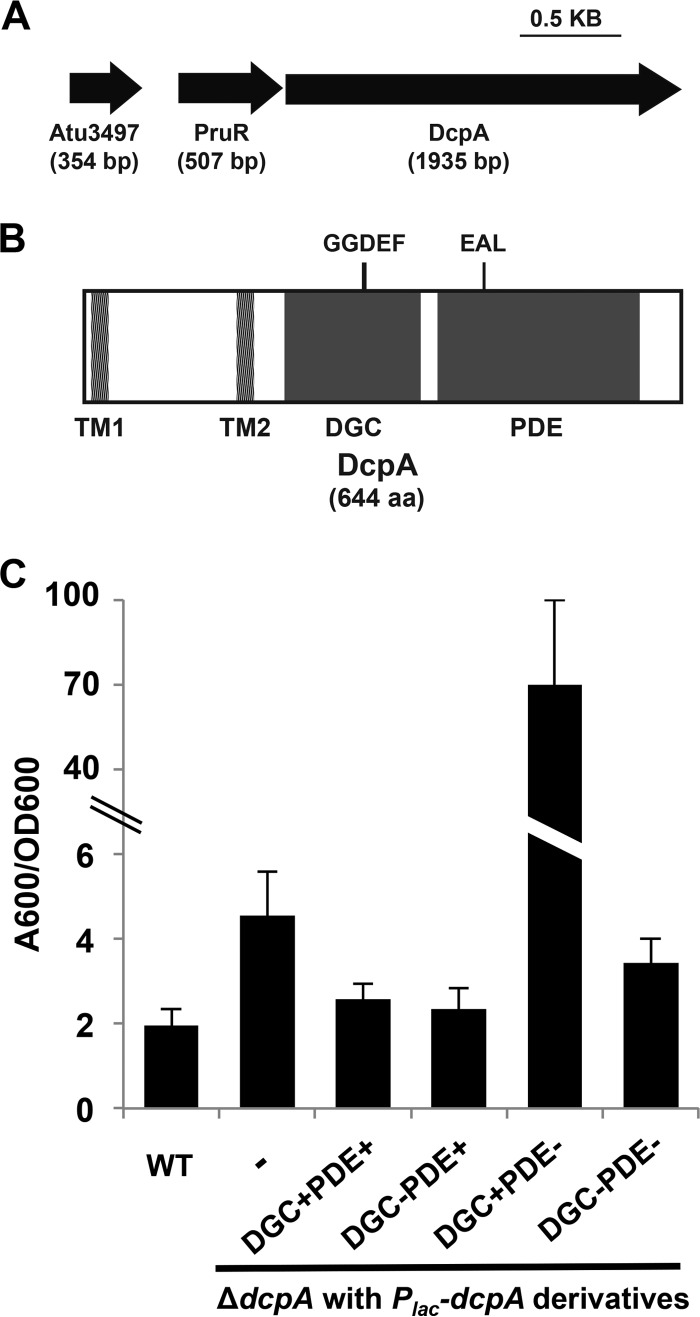
FIG 1 .
Complementation of increased biofilm formation in the A. tumefaciens dcpA mutant requires an intact DcpA EAL catalytic motif. (A) Diagram of PruR-DcpA genetic locus. Atu3497 is a conserved hypothetical protein with no annotated domains. (B) Protein topology of DcpA. Protein domains predicted by the BLAST database (NCBI) are shown. Domains are drawn to scale. TM, transmembrane; DGC, diguanylate cyclase; PDE, phosphodiesterase. (C) A. tumefaciens quantitative biofilm formation on PVC coverslips after 48 h of static growth at 28°C. Adherent biomass was quantified by staining with crystal violet (CV). CV absorbance was quantified by absorbance at 600 nm (A600). In parallel, the optical density at 600 nm (OD600) of planktonic culture was determined. CV absorbance was normalized to culture growth by calculating the A600/OD600 ratio. IPTG (400 µM) was added to all strains. The wild-type (WT) strain, ΔdcpA mutant with no plasmid inserted (-), and ΔdcpA mutant strain with Plac-dcpA derivatives are shown. A wild-type copy of dcpA provided on a plasmid expressed from the lacZ promoter (Plac-dcpA; DGC+PDE+) was introduced into a ΔdcpA mutant. The Plac-dcpA derivatives include a DcpA variant containing catalytic site GGDEF→GGDAF mutation (DGC−) and a DcpA variant containing catalytic site EAL→AAL mutation (PDE−). Values are results of three independent biological replicates consisting of three technical replicates each. The error bars show 1 standard deviation (SD).