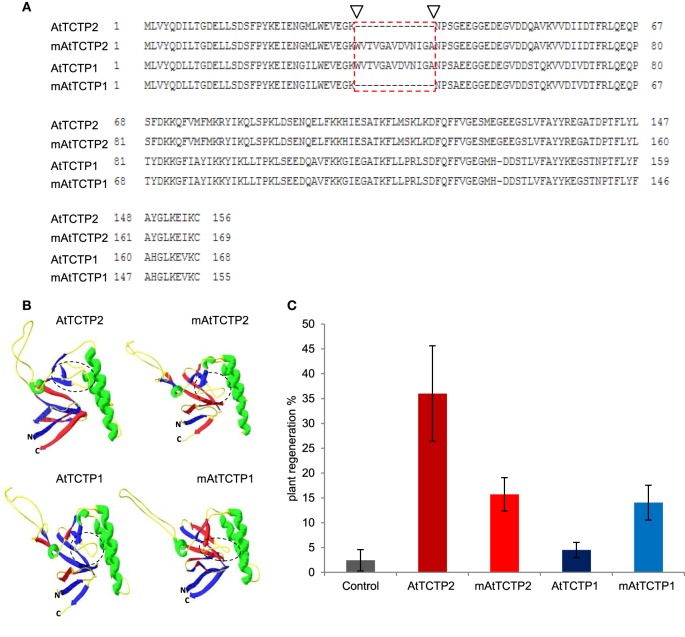
Figure 10.
Domain swapping between AtTCTP2 and AtTCTP1 alters their regeneration capacity. (A) Amino acid sequence alignment of AtTCTP2 and AtTCTP1 versions with ClustalX. The main difference between unmodified (normal/endogenous) protein versions is located in the 13 amino acid region shown by arrowheads and red-dashed rectangle; this region was swapped between AtTCTP1 and AtTCTP2 in order to generate the modified versions for both proteins. (B) Predictive three-dimensional structures of AtTCTP2 and AtTCTP1 (unmodified and modified versions) were modeled with the SWISS-MODEL automated protein structure homology-modeling server and processed using Deep View program (Swiss-Pdb-Viewer); dashed circles underline the region with more structural differences in all four versions. (C) Rate of plant regeneration is altered when both AtTCTP2 and AtTCTP1 are modified. Transformation of tobacco explants was performed with the four-overexpression versions (fused to GFP) previously mentioned. Three transformation events were performed with 30 explants per biological replicate (n ≈ 90), given as mean ± SE. Plant regeneration rate of AtTCTP2 was reduced significantly while that of AtTCTP1 increased in the modified versions (mAtTCTP2 and mAtTCTP1) relative to the unmodified ones (AtTCTP2 and AtTCTP1).