Figure 1.
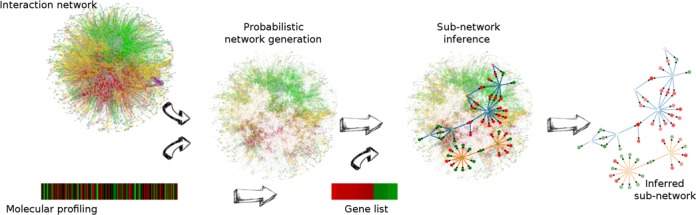
Overview of PheNetic, a web service for network-based interpretation of ‘omics’ data. The web service uses as input a genome wide interaction network for the organism of interest, a user generated molecular profiling data set and a gene list derived from these data. Interaction networks for a wide variety of organisms are readily available from the web server. Using the uploaded user-generated molecular data the interaction network is converted into a probabilistic network: edges receive a probability proportional to the levels measured for the terminal nodes in the molecular profiling data set. This probabilistic interaction network is used to infer the sub-network that best links the genes from the gene list. The inferred sub-network provides a trade-off between linking as many genes as possible from the gene list and selecting the least number of edges.
