Figure 1.
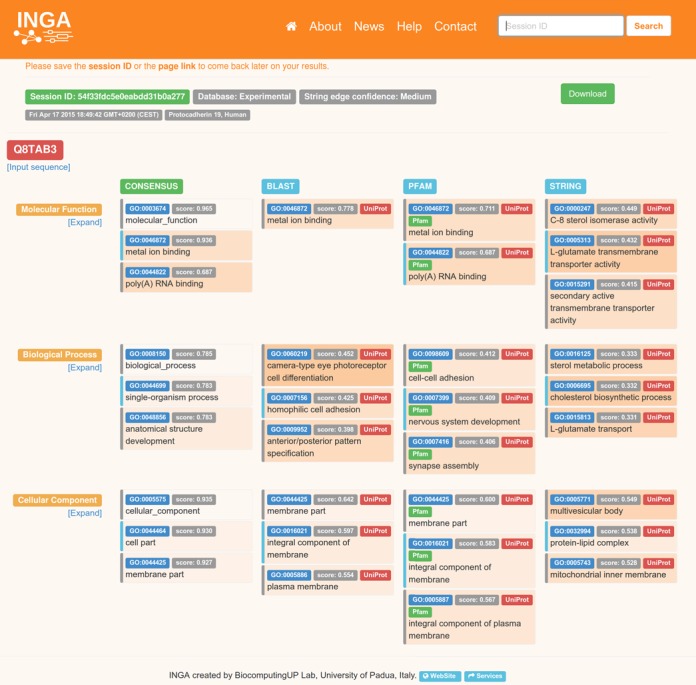
INGA results page for the human PCDH19 protein. The output contains a short header, with the session identifier, optional job title, download button as well as the UniProt accession code or title of the Fasta file used as input. The following three sections cover the GO sub-ontologies for molecular function, biological process and cellular component. Inside each section, the first column represents the consensus and is followed by BLAST, PFAM and STRING results. Each component result lists the GO term identifier, score and GO term description inside a box of variable background color. Darker background colors correspond to more informative (i.e. deeper) GO levels. Scores are in the range 0.0 (low) to 1.0 (high), with 1.0 reserved for curated GO terms for the query protein retrieved from UniProt. Where possible, UniProt and Pfam buttons open pop-up windows listing the UniProt accession numbers supporting the predicted GO term. An alternating gray or light blue stripe on the left side of the results box indicates whether consecutive entries belong to the same prediction or not. This is important, as multiple GO tracing the way from the prediction back to the ontology root can be shown. By default, only the top three results are shown for each GO sub-ontology and method. Clicking on ‘Expand’ below the GO sub-ontology name will expand the results section to cover all informative predictions for each method. Clicking on ‘Expand all’ will increase the number of visualized predictions further by listing all parent nodes of each prediction.
