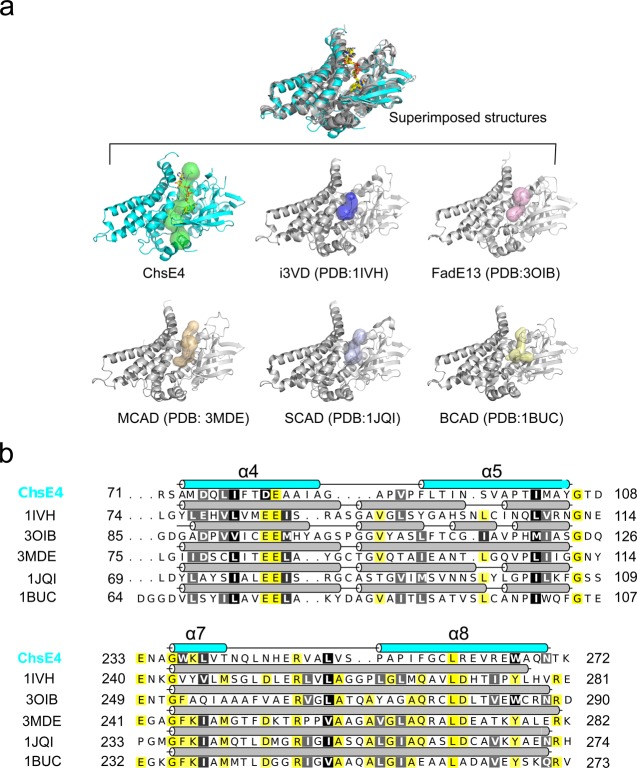
Figure 7.
Substrate binding site comparison across species. (a) The structures of ChsE4, human isovaleryl-CoA dehydrogenase (i3VD), pig medium chain acyl-CoA dehydrogenase (MCAD), Mycobacterium smegmatis FadE13, rat short chain acyl-CoA dehydrogenase (SCAD), and Megasphaera elsdenii butyryl-CoA dehydrogenase (BCAD) are superimposed, and their substrate binding sites have been identified by Caver.28 The substrate binding sites are shown as transparent surfaces. The RMSD value between ChsE4 and i3VD is 2.030 Å with 975 α-carbons aligned; the RMSD value between ChsE4 and 3OIB is 1.718 Å with 831 α-carbons aligned; the RMSD value between ChsE4 and 3MDE is 2.362 Å with 942 α-carbons aligned; the RMSD value between ChsE4 and 1JQI is 2.714 Å with 941 α-carbons aligned; the RMSD value between ChsE4 and 1BUC is 2.086 Å with 870 α-carbons aligned. (b) Secondary structure sequence alignment of ChsE4, i3VD, MCAD, SCAD, BCAD, and FadE13. ChsE4 is colored in cyan, and the other secondary structure cartoons are colored in gray. Yellow highlighted residues are identical; black and gray highlighted residues are very similar.