Figure 1.
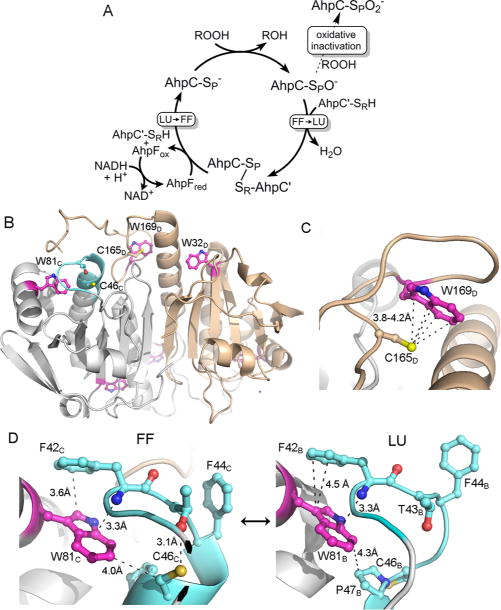
Catalytic cycle of 2-Cys peroxiredoxins and the location of the three Trp residues in Salmonella typhimurium AhpC. (A) The catalytic cycle (solid arrows) is shown along with the peroxidemediated inactivation pathway (dashed arrow). The conformation change necessary for Prx catalysis is shown as a transition from fully folded (FF) to locally unfolded (LU) enzyme after sulfenate formation at the peroxidatic Cys (shown as −SPO−), and the opposite transition that occurs upon reduction of the disulfide bond between SP and SR (the sulfur group of the resolving Cys) to regenerate the active enzyme. Oxidative inactivation by peroxide can also occur at the peroxidatic Cys (dashed arrow) to generate the cysteine sulfinate (shown as −SPO2−). (B) A single dimer [taken from a 1.8 Ắ resolution decameric crystal structure of reduced, wild-type AhpC (Protein Data Bank entry 4MA9)] is depicted with chain D colored tan and the partner subunit (chain C) colored gray, highlighting the location of Trp residues (magenta), Cys residues, and the regions that undergo structural changes (cyan) during catalysis (sulfur atoms colored yellow, nitrogen atoms blue, and oxygen atoms red). (C) Close-up of the region around C165 in the FF conformation. In the LU conformation, W169 is presumed to be disordered as no electron density is observed for it. (D) Close-ups of the region around W81 in the FF and LU conformations. Selected residues and atomic distances (dashed lines) in panels C and D are shown with the same color scheme as in panel B. Subscript letters after the residue numbers indicate the subunit of that residue (described in ref 30). Shown to the right in panel D is the LU conformation only of subunit B in Protein Data Bank entry 4MA9.
