Abstract
Foamy virus (FV) vectors are promising for hematopoietic stem cell (HSC) gene therapy but preclinical data on the clonal composition of FV vector transduced human repopulating cells is needed. Human CD34+ human cord blood cells were transduced with an FV vector encoding a methylguanine methyltransferase (MGMT)P140K transgene, transplanted into immunodeficient NOD/SCID IL2Rγnull (NSG) mice, and selected in vivo for gene-modified cells. The retroviral insertion site (RIS) profile of repopulating clones was examined using modified genomic sequencing PCR (MGS-PCR). We observed polyclonal repopulation with no evidence of clonal dominance even with the use of a strong internal spleen focus forming virus (SFFV) promoter known to be genotoxic. Our data supports the use of FV vectors with MGMTP140K for HSC gene therapy, but also suggests additional safety features should be developed and evaluated.
Keywords: Hematopoietic stem cells, gene therapy, genetic transduction, genetic vectors, foamy virus vectors, genotoxicity, chemoselection, methylguanine methyltransferase, MGMTP140K, NSG mouse
INTRODUCTION
Retroviral vectors derived from the foamy viruses (FVs) efficiently transduce hematopoietic stem cells (HSCs) and are promising for use in HSC gene therapy.1 One challenge for HSC gene therapy is that some diseases like hemoglobinopathies require high percentages of gene marked cells in order to achieve therapeutic correction. One strategy to overcome low marking is to engineer vectors with drug resistance genes that can be used to chemoselect gene-modified cells in vivo. Alkylating agents bis-chloroethylnitrosourea (BCNU) and temozolomide damage cells by forming toxic O6-guanine DNA adducts. These lesions are normally repaired by the gene product of cellular O6-methylguanine-DNA-methyltransferase (MGMT). O6-Benzylguanine (O6BG) efficiently inactivates the gene product of wild type MGMT,2 but not the MGMTP140K mutant.3
Mutant MGMT genes encoding O6BG resistant proteins have been included in retroviral vectors as chemoselectable markers for transduced cells.4, 5 While MGMT-mediated chemoselection can effectively increase the proportion of gene marked cells in a transplant recipient,5 in vivo selection pressures might also increase genotoxic risk and promote clonal dominance.6 Thus it’s important to evaluate the effect of chemoselection on clonality prior to the use of therapeutic vectors in the clinic.
The effect of MGMT-mediated chemoselection on clonality has been studied in detail for gammaretroviral (GV) and lentiviral (LV) vectors. In vivo selection studies examining clonality have been conducted in dogs (GV7–9; LV7, 9), in humanized mice (LV10), and in non-human primates (GV11; LV11, 12). Clonality data has also been published for chemoselected human cells transduced with an MGMTP140K expressing GV vector in a glioblastoma trial.13 MGMTP140K-mediated selection has been evaluated for FV vectors both in vitro14 and in animal models.14–16 However, to our knowledge, detailed clonality and integration site data for FV vectors in human repopulating cells after chemoselection in vivo has yet to be reported.
Here we examined clonality and retroviral insertion sites (RIS) of FV vectors in human SCID-repopulating cells after in vivo selection.
RESULTS AND DISCUSSION
Our goal was to investigate the genotoxicity of FV vectors in the context of in vivo selection in human hematopoietic cells. Gene modified SCID-repopulating cells were selected in vivo using O6BG and BCNU, which has the potential to increase genotoxic risk.6 For this study we used the strong spleen focus forming virus (SFFV) promoter, which has a high potential to contribute to vector genotoxicity by dysregulating host genes near integrated vector proviruses.17 This approach, with a genotoxic strong viral promoter and in vivo selection to enhance proliferation, can be viewed as a worst case scenario to explore the genotoxicity of FV vectors in human SCID-repopulating cells.
Xenotransplantation and in vivo selection of FV vector transduced SCID-repopulating cells
FV vector FV-SMPGW-KO (Figure 1a) was concentrated to 1×108 TU/mL and used to transduce human CD34+ cord blood cells, which were then transplanted into NOD/SCID IL2Rγnull (NSG) mice. Vector exposed cells were maintained in vitro in liquid culture and in semisolid methylcellulose media for colony forming unit (CFU) assay (Supplementary figure 1). Marking was 23.1% in liquid culture (Figure 1b) and 29% In the CFU assay, demonstrating efficient transduction of hematopoietic progenitors. Plating efficiency in the CFU assay was high (15%), indicating low vector cytotoxicity.
Figure 1.
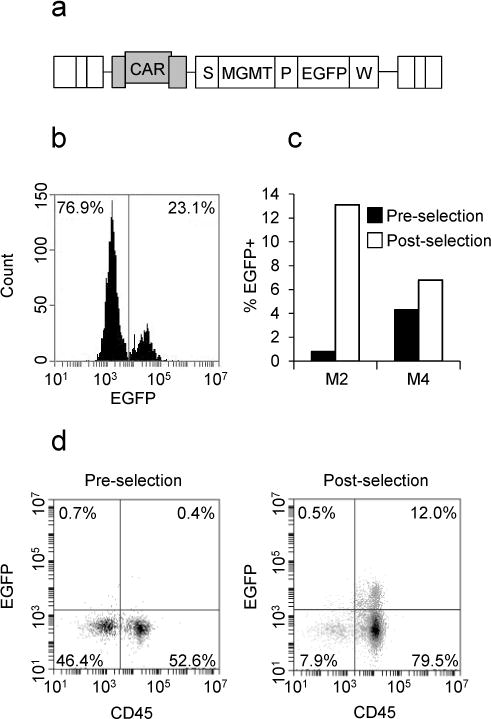
Vector design and marking. (a) FV vector FV-SMPGW-KO. Abbreviations: CAR, cis-acting region; EGFP, enhanced green fluorescent protein; MGMT, MGMTP140K; P, phosphoglycerate kinase promoter; S, spleen focus forming virus promoter; W, woochuck hepatitis virus post transcriptional regulatory element. (b) Marking in CD34+ human cord blood cells exposed to FV vector and maintained in liquid culture for 6 days. (c) Marking in CD45+ bone marrow cells of two mice which responded to selection. (d) Representative flow cytometry data.
During week 12 post-transplant, marking and engraftment were evaluated in bone marrow. Engraftment was 50% or greater in 3 of 4 animals, similar to what has been previously reported.18 Marking as a percentage of CD45+ cells was up to 6% (Supplementary table 1). Marking levels in vivo may have been negatively impacted by highly expressed MGMTP140K, as has been noted previously.19
To select for transduced cells, mice were treated with a chemotherapeutic regimen that selectively depletes cells not expressing MGMTP140K. During weeks 15–18 post-transplant, mice were treated 3 times with O6BG and BCNU. Mice were sacrificed during weeks 20–22 post-transplant and bone marrow and spleen were collected. Of four mice treated, we observed clear evidence of selection in two animals (>50% increase in EGFP+ human CD45+ cells) (Figure 1c–d, supplementary table 1). In mouse M2, marking increased by 16-fold, and in mouse M4 marking increased by more than 50% in response to selection. Selection in these mice was comparable to previously published studies in the humanized NSG mouse using GV and LV vectors, where EGFP increased by 50%–30 fold relative to preselection expression.10, 19–21 Mouse M1 had low engraftment and was not analyzed. Mouse M3 had high engraftment and low marking and did not respond to chemoselection. It is not clear to us why marking did not increase in this animal. However, it has been suggested that MGMTP140K expressed from the SFFV promoter might reduce the effectiveness of chemoselection.19, 22
Analysis of integration sites in FV vector transduced SCID-repopulating cells
We used modified genomic sequencing PCR (MGS-PCR),20 a modified linear amplification mediated PCR (LAM-PCR) protocol which omits initial linear amplification to improve the accuracy of clonal contribution data, to recover post-selection vector integration sites from bone marrow and spleen of both mice that responded to selection. Reads were processed using the Vector Integration Site Analysis (VISA) bioinformatics server21 to localize sites to the human genome. A total of 139 unique RIS were recovered (Figure 2). Bone marrow and spleen cell populations were polyclonal following selection. There was no evidence of clonal dominance in any of the tissues analyzed or in the combined set of all unique RIS (Figure 2).
Figure 2.
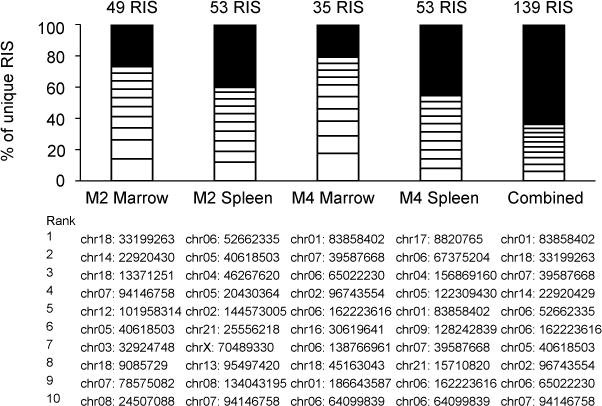
Clonality in bone marrow and spleen. RIS ranked 1–10 by capture frequency are shown as white rectangles. All others are shown as black rectangles. Locations are indicated for the ten most frequently captured RIS in each sample. Abbreviations: chr, chromosome; M, mouse; RIS, retroviral insertion site.
To determine whether RIS near particular gene classes were over-represented in our data, the nearest transcription start site (TSS) within a 100 kb window of each RIS was tabulated, and a gene ontology term enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID).22, 23 In previous studies of lentiviral and gammaretroviral RIS in primate repopulating cells, RIS were enriched near genes involved in growth and survival.24 Here, there was no significant enrichment in vivo in any of the ontology terms analyzed (Supplementary table 2).
We next compared our data to several successively larger lists of proto-oncogenes. These were, in order of list size: Leukemia (682 genes), the Retrovirus and Transposon Tagged Cancer Gene Database (RTCGD)25 and the Catalogue of Somatic Mutations in Cancer (COSMIC)26 (943 genes), and The Network of Cancer Genes 4.0 (NCG 4.0) (2048 genes).27, 28 For each of these lists, post-selection RIS were binned by distance to the nearest proto-oncogene TSS. RIS in each bin were then plotted as a percentage of unique RIS (Figure 3a–c). The distributions were similar between datasets and across oncogene lists. When all RIS in proto-oncogene transcripts or within 50 kb of proto-oncogene TSS were considered together, the distribution of in vivo RIS was significantly different (p < 0.05) from random for NCG 4.0 oncogenes (Figure 3d), but not for Leukemia or RTCGD + COSMIC oncogenes. When in vivo RIS were ranked by recovery frequency and divided into high, mid, and low thirds (Supplementary table 3), RIS in the highest third were significantly enriched in or near NCG 4.0 proto-oncogenes when compared to RIS in the lowest third (Figure 3d). Thus, although clonal dominance was not observed in this study, it is possible there is a subtle competitive growth advantage for cells with RIS in or near proto-oncogenes. In future studies the use of insulators and/or lineage-restricted promoters should be evaluated in FV vectors to determine if they can reduce or eliminate this effect.
Figure 3.
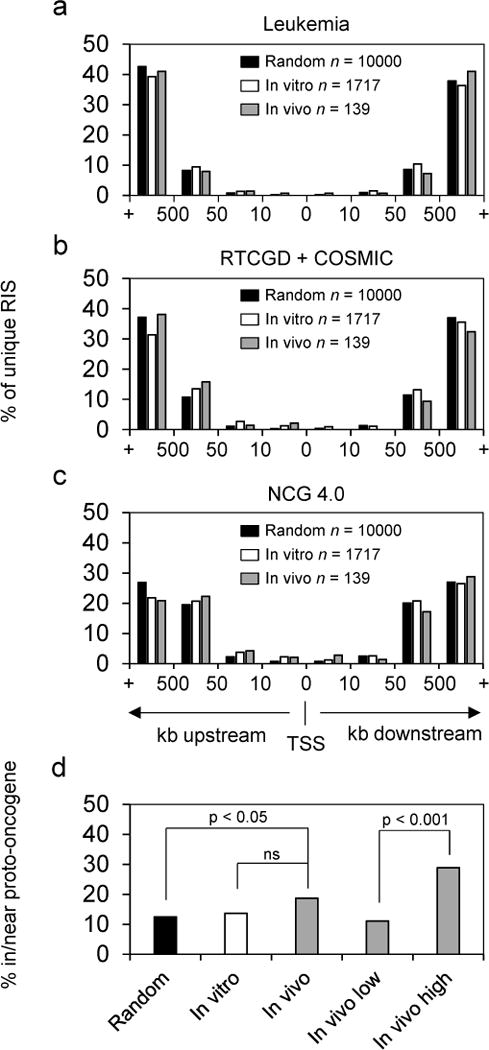
Proximity of RIS to TSS of proto-oncogenes in cancer gene lists. (a) Leukemia (n = 682) (b) Retrovirus and Transposon Tagged Cancer Gene Database (RTCGD) and the Catalogue of Somatic Mutations in Cancer (COSMIC) (n = 943). (c) Network of Cancer Genes 4.0 (NCG 4.0) (n = 2048). (d) Proportion of RIS within NCG 4.0 proto-oncogene transcripts or within 50 kb of a proto-oncogene TSS. In vivo low and in vivo high represent the lowest and highest thirds of RIS ranked by recovery frequency. Abbreviations: ns, not significant; RIS, retroviral insertion site; TSS, transcription start site.
Integration hotspots have been defined previously as groups of three or more RIS within 100 kb.29 There were no groups of three or more RIS within 100 kb in our data. The closest four RIS (Supplementary Figure 2) were near the genes LGSN, PHF3, EYS, and PTP4A1 on chromosome 6. LGSN encodes a pseudo-glutamine synthetase that has been associated with lung cancer.30 PHF3 encodes a candidate transcription factor that has been associated with glioblastoma.31 EYS encodes a protein with multiple epidermal growth factor-like domains that is localized to the retina.32 PTP4A1 (Alias PRL-1) encodes a member of a family of prenylated protein tyrosine phosphatases; signaling molecules which affect cell growth and cycling33–36 and are thought to be important in HSC self-renewal and proliferation.37 Given these functions, it is possible that dysregulation of PTP4A1 and/or these other nearby genes by integrated vector proviruses could provide a competitive growth advantage in repopulating cells. In future studies with FV vectors in human HSCs it may be useful to monitor integration at this locus.
In summary we have established polyclonal marking following MGMTP140K-mediated selection of human SCID-repopulating cells in vivo. Using an FV vector with a strong SFFV promoter known to be genotoxic, we observed polyclonal reconstitution without clonal dominance. Our preclinical data further establish the safety of FV vectors and support the use of FV MGMTP140K vectors.
MATERIALS AND METHODS
Vector production
Vector FV-SMPGW-KO contains a spleen focus forming virus (SFFV) promoter driving expression of MGMTP140K, a phosphoglycerate kinase (PGK) promoter driving expression of EGFP, and a woodchuck hepatitis virus post transcriptional regulatory element (WPRE) (Figure 1a). It also contains a bacterial origin of replication and kanamycin resistance gene.
Vectors were produced by transient transfection of vector plasmid and helper plasmids on HEK-293 cells using polyethylenimine as previously described,14 except that 14 μg of vector plasmid FV-SMPGW-KO, 4, 6, and 0.3 μg of FV helper plasmids pFVGagCO, pFVPolCO, pFVEnvCO, and 72.9 μL of 1 μg/μL polyethylenimine were used and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid was omitted. The FV helper plasmids were codon optimized to improve expression and to eliminate the potential for recombination. Vector-containing supernatant was passed through a 0.45 μm filter, concentrated 100-fold by ultracentrifugation at 23 °C, and frozen at −80 °C until use in serum-free media containing 5% DMSO. Before use, vector preparations were dialyzed using an Amicon™ Ultra 0.5 mL Centrifugal 50K Filter (UFC505024, Fisher Scientific, Waltham, MA, USA) to remove DMSO. Vector preparations were titered on HT1080 cells and EGFP-expressing cells were quantified using a BD Accuri C6 Flow Cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Transduction of CD34+ human cord blood cells and progenitor assay
Cryopreserved CD34+ human cord blood cells (Cat# 2C-101, Lonza Poietics, Basel, Switzerland) from a male donor were thawed, counted, and plated in a 12-well tissue culture treated plate at 2×105–1×106 cells/mL in prestimulation media (IMDM + 10% heat inactivated FBS with 5000U penicillin/streptomycin and cytokines: rhIL-3 (Cat# CYT-210), rhIL-6 (Cat# CYT-213), rhSCF (Cat# CYT-255), rhTPO (Cat# CYT-302), rhFlt-3 (Cat# CYT-331), rhG-CSF (Cat# CYT-220) (ProSpec-Tany TechnoGene Ltd., Rehovot, Israel), 100ng/mL each. Cells were incubated at 37 °C overnight. The following morning, a human fibronectin fragment coated 12-well suspension plate was prepared by coating wells with 2 μg/cm2 RetroNectin® Reagent (Cat# T100A, Takara Bio, Otsu, Shiga, Japan). Cells were counted and plated in three wells at 9×105 cells/well in prestimulation media. Cells were exposed to FV vector at an MOI of 10, or were mock transduced with prestimulation media. The final volume of each well was adjusted to 1 mL with prestimulation media and cells were incubated at 37 °C for 19 hours. Cells were washed and counted and an appropriate number of cells were removed for transplantation and progenitor assay. Remaining cells were maintained in liquid culture for 5 more days and then analyzed for EGFP expression by flow cytometry. Events were viewed on a live cell gate. One day after cells were exposed to vector, 2000 cells were plated in semisolid Methocult™ methylcellulose media (Cat# 04230, Stemcell Technologies, Vancouver, BC, Canada). Methocult™ was prepared according to manufacturer’s directions, and the following cytokines were added: 50 ng/mL rhSCF, 20 ng/mL rhIL-3, 20 ng/mL rhG-CSF, 20 ng/mL rhGM-CSF (Cat# CYT-089) (Prospec-Tany TechnoGene Ltd., Rehovot, Israel). Methocult™ plates were incubated at 37 °C. 11 days later, CFUs were counted and scored for EGFP expression using fluorescence microscopy.
Xenotransplantation in NSG mice
All animal procedures were reviewed and approved by the Washington State University Institutional Animal Care and Use Committee. NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, Cat# 005557, The Jackson Laboratory, Bar Harbor, ME, USA) were housed in sterile microisolator cages. The day before receiving transplants, six-week-old female NSG mice received 300 cGy total body irradiation. The day of transplant, cells were counted, then washed and resuspended in buffered saline. 1×105 cells were delivered by tail vein injection.
In vivo marking analysis
Bone marrow was collected from mice at 12 weeks post-transplant using a previously described non-lethal procedure.38 To create single cell suspensions, whole spleens were pressed through a fine wire mesh using a rubber policeman, passed through a 100 μm cell strainer (Cat# 352360, Corning, Painted Post, NY, USA), resuspended and incubated for five minutes at room temperature in 1× ACK lysis buffer (0.15M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA-Na2, filtered 0.22 μm) to remove RBCs. Cells were washed and resuspended in wash solution (DPBS + 0.1% BSA, filtered .22 μm), and strained through a 40 μm cell strainer (Cat# 352340, Corning, Painted Post, NY, USA). Crude marrow samples were passed through a 40 μm cell strainer, resuspended and incubated for five minutes at room temperature in 1× ACK lysis buffer, then washed and resuspended in wash solution. Single cell suspensions were counted using a haemocytometer and an appropriate number of cells were resuspended in staining buffer (DPBS + 1% BSA + 0.5% sodium azide, filtered .22 μm). For bone marrow cells, 5×105 mononuclear cells were used for each staining reaction. Following an Fc receptor blocking step, samples were stained for thirty minutes at 4 °C in the dark with APC-conjugated mouse anti-human CD45 (Clone HI30) and PerCP-conjugated rat anti-mouse CD45 (Clone 30-F11) (BD Biosciences, Franklin Lakes, NJ, USA) antibodies. Isotype controls were stained with APC-conjugated mouse IgG1, Κ (Clone MOPC-21) and PerCP-conjugated rat IgG2b, Κ (Clone A95-1) (BD Biosciences, Franklin Lakes, NJ, USA) antibodies. Cells were washed and then analyzed on a BD Accuri C6 flow cytometer.
In vivo selection of transduced SCID-repopulating cells
During weeks 15–18 post-transplant, mice were treated three times with 5 mg/kg O6BG (Cat# B2292), and 7.5 mg/kg BCNU (Cat# C0400) (Sigma-Aldrich, St. Louis, MO, USA) in order to select for transduced cells expressing MGMTP140K. O6BG was prepared at 1 mg/mL in a solution of 40% PEG-400 in DPBS and diluted to 0.2 mg/mL in DPBS before injection. BCNU was prepared at 2.14 mg/mL in a solution of 10% ethanol and DPBS and used undiluted. Timed doses were delivered by intraperitoneal injection. At each treatment, O6BG was delivered in two injections spaced fifteen minutes apart. BCNU was delivered in a single injection one hour after the first O6BG injection. Mice were monitored overnight following treatment.
Analysis of integration sites in FV vector transduced SCID-repopulating cells
Genomic DNA was extracted from single cell suspensions of bone marrow and spleen using the Gentra Puregene Cell Kit (Cat# 158388, Qiagen, Valencia, CA, USA), according to the manufacturer’s directions. Vector-genome junctions were sequenced using MGS-PCR as previously described.20 The Vector Integration Site Analysis (VISA) server21 (https://visa.pharmacy.wsu.edu/bioinformatics/) was used to process and map vector-genome junctions to the Genome Reference Consortium (GRC) build GRhg38 of the human genome,39 and to identify nearby genes and promoters. Sequences that could not be confidently localized were removed from the dataset before analysis. VISA was used to generate a set of 10 000 random RIS in silico21 for comparison with the in vivo dataset. For comparison to in vitro RIS, a previously published data set29 was used (Supplementary table 4).
Clonality was assessed by ranking RIS by recovery frequency and normalizing to the total (Non-unique, localizable) number of RIS reads recovered for each sample. For the combined in vivo dataset, recovery frequencies were normalized to the total number of RIS reads recovered for all samples.
Enrichment analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 6.7 (http://david.abcc.ncifcrf.gov). The database was queried with the list of Refseq genes with nearest TSS within a 100 kb window of each RIS. Functional annotation charts were generated using each of the following GO terms: Biological Process (BP = GOTERM_BP_ALL), Molecular Function (MF = GOTERM_MF_ALL), and Protein Information Resource (PIR = SP_PIR_KEYWORDS). For each analysis, EASE threshold was set to 1 and count threshold was set to 3. Significance was assessed using Fisher’s exact test with corrections for multiple comparisons (Holm-Bonferroni method).40 Results were considered significant at p < 0.05.
RIS within or near proto-oncogenes were identified using PERL scripts querying a set of 682 leukemia genes from NCG 4.0,27, 28 (http://www.bioinformatics.org/legend/leuk_db.htm#g1) and from the Bushman lab (http://www.bushmanlab.org/links/genelists), a set of 943 proto-oncogenes from RTCGD25 and COSMIC,26 or a set of 2048 proto-oncogenes from NCG 4.0. An RIS was considered to be within or near an oncogene when located within an oncogene transcript and/or when an oncogene TSS was within a 100 kb window of the RIS. For comparisons of the frequency of RIS within or near an oncogene, the χ2 test was used. Results were considered significant at p < 0.05.
Hotspots were defined as regions with three or more RIS within a 100 kb window, as has been previously described.29 The nearest four RIS were also identified and plotted together in the UCSC genome browser.41
Supplementary Material
Acknowledgments
This research was supported in part by NIH grants AI097100 (HPK, DJR and GDT), and AI102672 (GDT). We thank Diana Browning for advice and assistance with flow cytometry.
Footnotes
CONFLICT OF INTEREST
The authors have no competing financial interests in relation to the work described.
Supplementary information is available at Gene Therapy’s website.
References
- 1.Trobridge GD. Foamy virus vectors for gene transfer. Expert Opin Biol Ther. 2009;9(11):1427–36. doi: 10.1517/14712590903246388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dolan ME, Moschel RC, Pegg AE. Depletion of mammalian O6-alkylguanine-DNA alkyltransferase activity by O6-benzylguanine provides a means to evaluate the role of this protein in protection against carcinogenic and therapeutic alkylating agents. Proceedings of the National Academy of Sciences. 1990;87(14):5368–5372. doi: 10.1073/pnas.87.14.5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu-Welliver M, Kanugula S, Pegg AE. Isolation of human O6-alkylguanine-DNA alkyltransferase mutants highly resistant to inactivation by O6-benzylguanine. Cancer research. 1998;58(9):1936–45. [PubMed] [Google Scholar]
- 4.Reese JS, Koc ON, Lee KM, Liu L, Allay JA, Phillips WP, Jr, et al. Retroviral transduction of a mutant methylguanine DNA methyltransferase gene into human CD34 cells confers resistance to O6-benzylguanine plus 1,3-bis(2-chloroethyl)-1-nitrosourea. Proc Natl Acad Sci U S A. 1996;93(24):14088–93. doi: 10.1073/pnas.93.24.14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis BM, Reese JS, Koc ON, Lee K, Schupp JE, Gerson SL. Selection for G156A O6-methylguanine DNA methyltransferase gene-transduced hematopoietic progenitors and protection from lethality in mice treated with O6-benzylguanine and 1,3-bis(2-chloroethyl)-1-nitrosourea. Cancer Res. 1997;57(22):5093–9. [PubMed] [Google Scholar]
- 6.Milsom MD, Williams DA. Live and let die: in vivo selection of gene-modified hematopoietic stem cells via MGMT-mediated chemoprotection. DNA Repair (Amst) 2007;6(8):1210–21. doi: 10.1016/j.dnarep.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neff T, Horn PA, Peterson LJ, Thomasson BM, Thompson J, Williams DA, et al. Methylguanine methyltransferase-mediated in vivo selection and chemoprotection of allogeneic stem cells in a large-animal model. J Clin Invest. 2003;112(10):1581–8. doi: 10.1172/JCI18782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neff T, Beard BC, Peterson LJ, Anandakumar P, Thompson J, Kiem HP. Polyclonal chemoprotection against temozolomide in a large-animal model of drug resistance gene therapy. Blood. 2005;105(3):997–1002. doi: 10.1182/blood-2004-08-3169. [DOI] [PubMed] [Google Scholar]
- 9.Beard BC, Sud R, Keyser KA, Ironside C, Neff T, Gerull S, et al. Long-term polyclonal and multilineage engraftment of methylguanine methyltransferase P140K gene-modified dog hematopoietic cells in primary and secondary recipients. Blood. 2009;113(21):5094–103. doi: 10.1182/blood-2008-09-176412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phaltane R, Haemmerle R, Rothe M, Modlich U, Moritz T. Efficiency and safety of O(6)-methylguanine DNA methyltransferase (MGMT(P140K))-mediated in vivo selection in a humanized mouse model. Hum Gene Ther. 2014;25(2):144–55. doi: 10.1089/hum.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beard BC, Trobridge GD, Ironside C, McCune JS, Adair JE, Kiem HP. Efficient and stable MGMT-mediated selection of long-term repopulating stem cells in nonhuman primates. J Clin Invest. 2010;120(7):2345–54. doi: 10.1172/JCI40767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larochelle A, Choi U, Shou Y, Naumann N, Loktionova NA, Clevenger JR, et al. In vivo selection of hematopoietic progenitor cells and temozolomide dose intensification in rhesus macaques through lentiviral transduction with a drug resistance gene. J Clin Invest. 2009;119(7):1952–63. doi: 10.1172/JCI37506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adair JE, Beard BC, Trobridge GD, Neff T, Rockhill JK, Silbergeld DL, et al. Extended Survival of Glioblastoma Patients After Chemoprotective HSC Gene Therapy. Sci Transl Med. 2012;4(133):133ra57. doi: 10.1126/scitranslmed.3003425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiem HP, Wu RA, Sun G, von Laer D, Rossi JJ, Trobridge GD. Foamy combinatorial anti-HIV vectors with MGMTP140K potently inhibit HIV-1 and SHIV replication and mediate selection in vivo. Gene Ther. 2010;17(1):37–49. doi: 10.1038/gt.2009.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trobridge GD, Beard BC, Wu RA, Ironside C, Malik P, Kiem HP. Stem cell selection in vivo using foamy vectors cures canine pyruvate kinase deficiency. PloS one. 2012;7(9):e45173. doi: 10.1371/journal.pone.0045173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai S, Ernstberger A, Wang H, Bailey BJ, Hartwell JR, Sinn AL, et al. In vivo selection of hematopoietic stem cells transduced at a low multiplicity-of-infection with a foamy viral MGMT(P140K) vector. Exp Hematol. 2008;36(3):283–92. doi: 10.1016/j.exphem.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montini E, Cesana D, Schmidt M, Sanvito F, Bartholomae CC, Ranzani M, et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J Clin Invest. 2009;119(4):964–75. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nasimuzzaman M, Kim Y-S, Wang Y-D, Persons DA. High-titer foamy virus vector transduction and integration sites of human CD34+ cell–derived SCID-repopulating cells. Molecular Therapy — Methods & Clinical Development. 2014;1:14020. doi: 10.1038/mtm.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milsom MD, Jerabek-Willemsen M, Harris CE, Schambach A, Broun E, Bailey J, et al. Reciprocal relationship between O6-methylguanine-DNA methyltransferase P140K expression level and chemoprotection of hematopoietic stem cells. Cancer Res. 2008;68(15):6171–80. doi: 10.1158/0008-5472.CAN-08-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai S, Wang H, Bailey B, Hartwell JR, Silver JM, Juliar BE, et al. Differential Secondary Reconstitution of In Vivo-Selected Human SCID-Repopulating Cells in NOD/SCID versus NOD/SCID/gamma chain Mice. Bone Marrow Res. 2011;2011:252953. doi: 10.1155/2011/252953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung J, Scherer LJ, Gu A, Gardner AM, Torres-Coronado M, Epps EW, et al. Optimized lentiviral vectors for HIV gene therapy: multiplexed expression of small RNAs and inclusion of MGMT(P140K) drug resistance gene. Mol Ther. 2014;22(5):952–63. doi: 10.1038/mt.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schambach A, Baum C. Vector design for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells. DNA Repair (Amst) 2007;6(8):1187–96. doi: 10.1016/j.dnarep.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beard BC, Adair JE, Trobridge GD, Kiem HP. High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods in molecular biology (Clifton, NJ. 2014;1185:321–44. doi: 10.1007/978-1-4939-1133-2_22. [DOI] [PubMed] [Google Scholar]
- 24.Hocum JD, Battrell LR, Maynard R, Adair JE, Beard BC, Kiem H-P, et al. VISA - Vector Integration Site Analysis Server: A Web-Based Server To Rapidly Identify Retroviral Integration Sites From Next-Generation Sequencing. Molecular Therapy. 2014;22(Suppl 1):S84. doi: 10.1186/s12859-015-0653-6. (abstract 220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beard BC, Dickerson D, Beebe K, Gooch C, Fletcher J, Okbinoglu T, et al. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol Ther. 2007;15(7):1356–65. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- 28.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic acids research. 2004;32:D523–7. doi: 10.1093/nar/gkh013. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91(2):355–8. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rambaldi D, Giorgi FM, Capuani F, Ciliberto A, Ciccarelli FD. Low duplicability and network fragility of cancer genes. Trends Genet. 2008;24(9):427–30. doi: 10.1016/j.tig.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 31.An O, Pendino V, D’Antonio M, Ratti E, Gentilini M, Ciccarelli FD. NCG 4.0: the network of cancer genes in the era of massive mutational screenings of cancer genomes. Database: the journal of biological databases and curation. 2014;2014:bau015. doi: 10.1093/database/bau015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trobridge GD, Miller DG, Jacobs MA, Allen JM, Kiem HP, Kaul R, et al. Foamy virus vector integration sites in normal human cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(5):1498–503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakatsugawa M, Hirohashi Y, Torigoe T, Asanuma H, Takahashi A, Inoda S, et al. Novel spliced form of a lens protein as a novel lung cancer antigen, Lengsin splicing variant 4. Cancer Sci. 2009;100(8):1485–93. doi: 10.1111/j.1349-7006.2009.01187.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Struss AK, Romeike BF, Munnia A, Nastainczyk W, Steudel WI, Konig J, et al. PHF3-specific antibody responses in over 60% of patients with glioblastoma multiforme. Oncogene. 2001;20(31):4107–14. doi: 10.1038/sj.onc.1204552. [DOI] [PubMed] [Google Scholar]
- 35.Collin RW, Littink KW, Klevering BJ, van den Born LI, Koenekoop RK, Zonneveld MN, et al. Identification of a 2 Mb Human Ortholog of Drosophila eyes shut/spacemaker that Is Mutated in Patients with Retinitis Pigmentosa. Am J Hum Genet. 2008;83(5):594–603. doi: 10.1016/j.ajhg.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diamond RH, Cressman DE, Laz TM, Abrams CS, Taub R. PRL-1, a unique nuclear protein tyrosine phosphatase, affects cell growth. Mol Cell Biol. 1994;14(6):3752–62. doi: 10.1128/mcb.14.6.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cates CA, Michael RL, Stayrook KR, Harvey KA, Burke YD, Randall SK, et al. Prenylation of oncogenic human PTP(CAAX) protein tyrosine phosphatases. Cancer Lett. 1996;110(1–2):49–55. doi: 10.1016/s0304-3835(96)04459-x. [DOI] [PubMed] [Google Scholar]
- 38.Werner SR, Lee PA, DeCamp MW, Crowell DN, Randall SK, Crowell PL. Enhanced cell cycle progression and down regulation of p21(Cip1/Waf1) by PRL tyrosine phosphatases. Cancer Lett. 2003;202(2):201–11. doi: 10.1016/s0304-3835(03)00517-2. [DOI] [PubMed] [Google Scholar]
- 39.Liang F, Liang J, Wang WQ, Sun JP, Udho E, Zhang ZY. PRL3 promotes cell invasion and proliferation by down-regulation of Csk leading to Src activation. J Biol Chem. 2007;282(8):5413–9. doi: 10.1074/jbc.M608940200. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi M, Bai Y, Dong Y, Yu H, Chen S, Gao R, et al. PRL2/PTP4A2 phosphatase is important for hematopoietic stem cell self-renewal. Stem Cells. 2014;32(7):1956–67. doi: 10.1002/stem.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verlinden SF, van Es HH, van Bekkum DW. Serial bone marrow sampling for long-term follow up of human hematopoiesis in NOD/SCID mice. Exp Hematol. 1998;26(7):627–30. [PubMed] [Google Scholar]
- 42.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 43.Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scandinavian Journal of Statistics. 1979;6(2):65–70. [Google Scholar]
- 44.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


