Fig. 4.
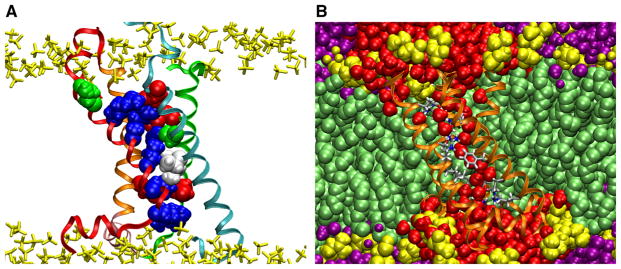
VSD structure and solvation. Snapshots from a 12 μs all-atom simulation of the VSD of the KV1.2 paddle-chimera channel (Long et al. 2007) in a lipid bilayer in excess water. a The VSD architecture houses two clusters of basic and acidic side chains separated by two hydrophobic residues. The residues determined by Palovcak et al. (2014) to be evolutionary “constrained” are shown as filled-spheres (basic, blue; acidic, red; polar, green; hydrophobic, white. See Fig. 3 for detailed sequence). The VSD is shown in ribbon representation with the same color scheme as in Fig. 2. The lipid phosphate groups are shown in yellow. b Solvation of the VSD by the membrane. Waters penetrate deeply into the VSD crevices. The VSD is shown in ribbon representation (orange) with the conserved residues in licorice representation colored by atom name. Waters within the first two coordination shells of the VSD are shown in red. Other waters are in purple. Lipids are shown as filled-spheres with the headgroups in yellow (Color figure online)
