We show that ATE1-encoded Arg-tRNA transferase (R-transferase) of the N-end rule pathway mediates N-terminal arginylation of multiple endoplasmic reticulum (ER)-residing chaperones, leading to their cytosolic relocalization and turnover. N-terminal arginylation of BiP/GRP78, protein disulfide isomerase (PDI), and calreticulin (CRT) is coinduced with autophagy during innate immune responses to cytosolic foreign DNA or proteasomal inhibition, associated with increased ubiquitination. Arginylated BiP (R-BiP) is induced by and associated with cytosolic misfolded proteins destined to p62/sequestosome-1/SQSTM1 bodies. R-BiP binds the autophagic adaptor p62 through the interaction of its N-terminal Arg (Nt-Arg) with p62 ZZ domain. This allosterically induces self-oligomerization and aggregation of p62 and increases p62 interaction with LC3, leading to p62 targeting to autophagosomes and selective lysosomal codegradation of R-BiP and p62 together with associated cargoes. In this autophagic mechanism, Nt-Arg functions as a delivery determinant, a degron and an activating ligand. Bioinformatics analysis predicts that many ER residents utilize arginylation to regulate non-ER processes.
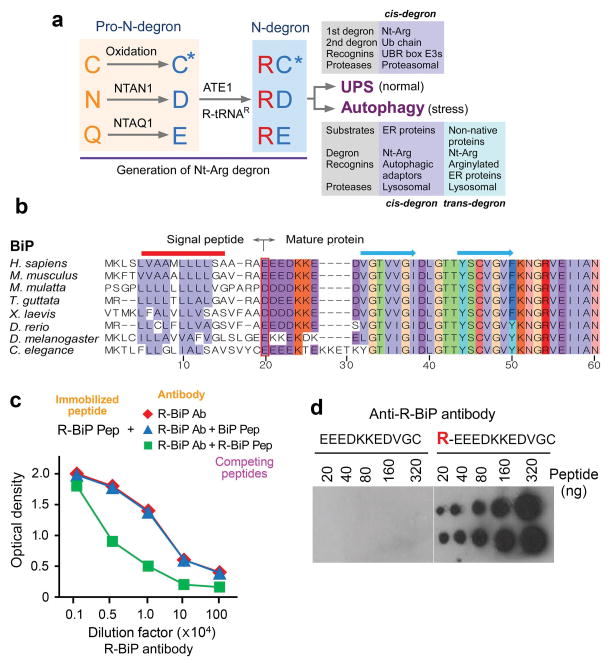
Substrate selectivity in proteolysis is governed by the interaction between degradation signals (degrons) and recognition components (recognins)1–2. A well characterized class of degrons in the Ub-proteasome system (UPS) is defined by the N-end rule pathway in which a destabilizing N-terminal residue functions as a determinant of a class of degrons, called N-degrons1,3,4,6. N-degrons are recognized and bound by N-recognins, such as those containing UBR boxes7–10, leading to proteolysis by the proteasome11–12 (Fig. 1a). A functional N-degron is composed of a destabilizing N-terminal residue as a binding ligand, an internal lysine (Lys) residue as a site of polyubiquitination, and an appropriate secondary/tertiary structure5,7,13–14. In mammals, destabilizing N-terminal residues include Arg, Lys, His (type 1; positively charged), Phe, Tyr, Trp, Leu, and Ile (type 2; bulky hydrophobic)15,16. These degradation determinants can be generated through endoproteolytic cleavages followed by post-translational modifications (PTMs)8–10, such as N-terminal arginylation by ATE1-encoded Arg-tRNA transferases (R-transferases; EC 2.3.2) that transfer L-Arg from Arg-tRNAArg to protein N-termini17,18 (Fig. 1a). In eukaryotes, Nt-aspartate (Asp) and Nt-glutamate (Glu) are the acceptors of N-terminal arginylation17,18. Nt-asparagine (Asn), Nt-glutamine (Gln), and Nt-cysteine (Cys) can also be arginylated following deamidation (of Nt-Asn and Nt-Gln)3,19,20 or oxidation (of Nt-Cys)18,19 (Fig. 1a). ATE1-knockout mouse embryos die with cardiovascular defects18,21,22. Recent studies have shown that Nt-methionine (Nt-Met)23 and N-terminally acetylated residues24 can also function as N-degrons.
Figure 1.
Bioinformatic analysis of the ER N-end rule pathway, and the generation of antibodies to the arginylated form of the ER chaperone BiP. (a) A summary of this study that describes a dual role for the Nt-Arg residue as a degron for both the UPS and autophagy. The Nt-Arg residue generated through the N-end rule pathway is known to act as a degron for selective proteolysis by the UPS. In the Ub-dependent N-end rule pathway, specific recognition components, called N-recognins, recognize and bind the primary degron Nt-Arg and mediate ubiquitination for targeting to the proteasome. The results of this study show that N-terminal arginylation of ER-residing chaperons generates a cis-acting delivery determinant and degron for lysosomal degradation along with associated cargoes. In this autophagic proteolysis, Nt-Arg of BiP and other ER proteins is recognized by a recognin, the autophagic adaptor p62. The binding of Nt-Arg to p62 activates p62, leading to the delivery of p62, R-BiP, and its cargoes to autophagosomes. In definition, Nt-Arg also acts as a trans-acting degron for its cargoes. (b) A sequence alignment of the N-terminal regions of human BiP and its sequelogs. The red box indicates the N-terminal residues (P1′ sites) of mature BiP proteins from various species. Shown above are secondary structures (solid line, αhelices; arrows, βstrands). (c) Generation of anti-R-BiP DH1 antibody. Shown is a peptide binding/competition assay. An 11-mer R-BiP peptide (R-BiP-peptide) corresponding to the N-terminal region of R-BiP was immobilized on a 96-well plate, followed by incubation with serially diluted anti-R-BiP antibody and, subsequently, anti-goat secondary antibody conjugated with horseradish peroxide. The amounts of R-BiP antibody that bound to immobilized R-BiP peptide were determined based on O.D. value at 450 nm of secondary antibody. As a control, BiP-peptide, a 10-mer peptide corresponding to the N-terminal region of unarginylated BiP, was used. (d) A dot blotting analysis of R-BiP antibody using the peptides corresponding to the N-terminal region of unarginylated (left) or arginylated (right) BiP.
Whereas PTM-generated degrons have been well characterized in the UPS1–7,25,26, little is known about the role of such degrons in proteolysis through the autophagy-lysosome system (hereafter called autophagy). Misfolded proteins that escape UPS-dependent quality control, including monomeric and oligomeric aggregates, may impair proteasomal and other cellular functions and, thus, are redirected to macroautophagy wherein cellular cargoes are segregated by autophagosomes for lysosomal degradation27–31. In macroautophagy, p62 selectively delivers Ub-conjugated cargoes to autophagosomes through self-aggregation and the interaction with LC332–33. One outstanding question in macroautophagy is how cargoes are selectively recognized by p62 and delivered to autophagosomes35,36.
Proteomic analysis of the N-end rule pathway for ER-residing proteins
Approximately one third of the human proteome is destined to the Golgi secretory pathway37,38. The signal peptides of ER clients and residents are cotranslationally and cotranslocationally cleaved off by the signal peptide peptidase, exposing new N-terminal residues on mature peptides39. Translocated polypeptides undergo folding and other PTMs through the assistance of folding factors such as BiP, GRP94, CRT, PDI, and ERdJ540. While these folding factors are known to be metabolically stable and function in the lumen lacking proteolytic machinery, they are also present in various non-ER compartments41,42. We hypothesized that these non-ER species acquire PTMs to support their activities, locations, and turnover. To characterize the N-end rule pathway for ER-residing proteins, we analyzed N-terminal residues of 498 ER-targeted proteins listed in the Human ER Aperçu (Hera) database43. Approximately 27% acquired destabilizing N-terminal residues (Supplementary Tables 1–4). In this study, we focused on two arginylation substrates (Asp and Glu) and three N-terminal residues (Asn, Gln, and Cys) whose PTMs confer arginylation through deamidation/arginylation (Asn and Gln) or oxidation/arginylation (Cys) (Fig. 1a). This arginylation-permissive group represented 9% (43 out of 498), amongst which 25 bore Asp or Glu (Supplementary Table 1). Of note, 11 retained arginylation permissiveness throughout evolution, including major Ca++-binding molecular chaperones (BiP, GRP94, and CRT) and oxidoreductases (PDI and ERdJ5) (Fig. 1b; Supplementary Fig. 1 and Table 3). All three proteins (BiP, PDI, and CRT) that we further characterized are shown to be N-terminally arginylated, indicating that many ER proteins may utilize similar N-terminal PTMs to participate in non-ER processes.
BiP is N-terminally arginylated by ATE1, and R-BiP functions in the cytosol
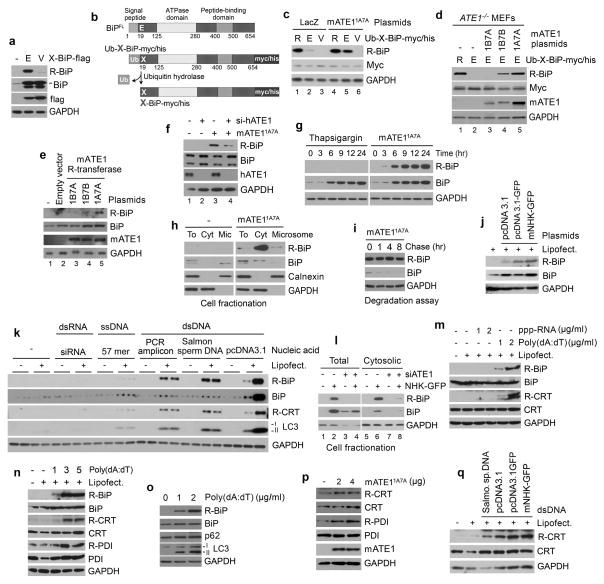
The arginylation-permissive P1′ residue, Nt-Glu19, of human BiP is evolutionarily conserved across various species (Fig. 1b). In agreement with this evolutionary pattern, an earlier mass spectrometric analysis of proteins separated on 2D-SDS PAGE identified arginylated forms of BiP and PDI44. We raised R-BiP DH1 antibody that exclusively recognizes R-BiP using the peptide REEEDKKEDVGC. The specificity was confirmed using immunoblotting, dot blotting, and peptide binding/competition assays (Fig. 1c, d and Supplementary Fig. 2). R-BiP was not readily detected in various cell lines. By overexpressing recombinant BiP, we confirmed that BiP was arginylated at Nt-Glu19 and that its arginylation was abolished by the Glu19-to-Val mutation (Fig. 2a). To further characterize BiP arginylation, we employed the Ub fusion technique8 wherein Ub-Glu19-BiP was cotranslationally cleaved by deubiquitination enzymes into Ub and Glu19-BiP (Fig. 2b). The arginylated mutant, Arg-Glu19-BiP, produced a stronger R-BiP signal compared with Glu19-BiP (Fig. 2c, lanes 1 vs. 2) in a manner independent of ATE1 function (Fig. 2d, lanes 1 vs. 2). Overexpressing ATE11A7A, an isoform containing alternative axons 1A and 7A, promoted N-terminal arginylation of recombinant Glu19-BiP in HEK293 cells (Fig. 2c, lanes 5 vs. 2) and ATE1−/− mouse embryonic fibroblasts (MEFs) (Fig. 2d, lanes 5 vs. 2) as well as endogenous BiP in HeLa cells (Fig. 2e, lanes 5 vs. 2). A moderate activity was also detected with ATE11B7B (Fig. 2d, lanes 4 vs. 2; 2e, lanes 4 vs. 2). ATE1-knockdown inhibited N-terminal arginylation of endogenous BiP (Fig. 2f, lanes 4 vs. 3). The ER stressor thapsigargin, an inhibitor of sarco/endoplasmic reticulum Ca2+ ATPase, did not readily induce BiP arginylation compared with overexpressed ATE11A7A (Fig. 2g), suggesting that R-BiP functions outside the ER. Indeed, R-BiP was retrieved from the cytosol (Fig. 2h) and excluded from the ER as determined by KDEL immunostaining (Supplementary Fig. 3). Consistent with an apparent lumen-to-cytosol flux of BiP, BiP was short-lived under translational inhibition (Fig. 2i).
Figure 2.
ATE1-dependent N-terminal arginylation of multiple ER-residing proteins is induced by cytosolic foreign dsDNA. (a) BiP is N-terminally arginylated at Nt-Glu19. HeLa cells were transfected with a plasmid expressing X-BiP-flag wherein the flag epitope was inserted between BiP protein body and the C-terminal KDEL sequence. (b) Schematic diagrams of full length BiP in comparison with Ub-X-BiP-myc/his in which Ub is C-terminally conjugated with N-terminal X residue (X= Glu, Arg-Glu, Val) of BiP-myc/his. In the Ub fusion technique, Ub-X-BiP-myc/his is cotranslationally cleaved by Ub hydrolases at the Ub-BiP junction, producing Ub and X-BiP-myc/his. (c) ATE11A7A promotes N-terminal arginylation of Glu19-BiP-myc/his in HEK293 cells cotranslationally generated from Ub-X-BiP-myc/his (X= Arg-Glu19, Glu19, or Val19). (d) N-terminal arginylation of X-BiP-myc/his (X= Arg-Glu19 or Glu19) was measured in ATE1−/− MEFs transiently expressing an ATE1 isoform (ATE11B7A, ATE11B7B, or ATE11A7A) which contains either of alternatively spliced exons (1A, 1B, 7A, and 7B). (e) N-terminal arginylation of endogenous BiP was measured in HeLa cells expressing an ATE1 isoform. (f) ATE1-knockdown inhibits the N-terminal arginylation of BiP in HeLa cells expressing ATE11A7A. (g) N-terminal arginylation of endogenous BiP is not induced by thapsigargin (200 nM) but by overexpressing ATE11A7A in HeLa cells. (h) Cell fractionation assay shows that R-BiP localizes to the cytosol. To, total extracts; Cyt, cytosolic fraction; Mic, microsomal fraction. (i) Lumenal BiP is short-lived, and endogenous R-BiP generated by ATE11A7A exhibits a longer half-life. HeLa cells expressing ATE11A7A were treated with 10 μg/ml cycloheximide, followed by time-course immunoblotting. (j–k) N-terminal arginylation of BiP and CRT is induced by the transfection of dsDNAs. Cells were incubated with various types of nucleic acids in the presence or absence of Lipofectamine LTX. Salmo. sp. DNA, Salmon sperm DNA. (l) DNA-induced R-BiP is cytosolic and down-regulated by ATE1-knockdown. HeLa cells were co-transfected with ATE1 siRNA #204 and a plasmid expressing NHK-GFP, followed by fractionation of cytosolic proteins. (m) N-terminal arginylation of BiP and CRT is induced by poly(dA:dT) dsDNA but not by 5′ppp-dsRNA. (n–q) N-terminal arginylation of ER proteins is triggered by poly(dA:dT) (n), coinduced with autophagy in poly(dA:dT)-treated HeLa cells (o), facilitated by overexpressing ATE11A7A (p), and induced by the transfection of various dsDNAs (q). mNHK-GFP, GFP-fused mouse α1-antitrypsin null Hong-Kong.
N-terminal arginylation of BiP is induced by cytosolic foreign DNA
We found that N-terminal arginylation of BiP was induced by transient transfection of various dsDNAs, including plasmid DNA and PCR-amplified DNA fragments (Fig. 2j, k). RNA interference assays showed that DNA-induced R-BiP was generated by ATE1 (Fig. 2l, lanes 4 vs. 2 and 8 vs. 6) and accumulated in the cytosol (Fig. 2l, lane 6). Extracellular dsDNA did not show such activity (Fig. 2k). Comparative assays (Fig. 2k, 2m and Supplementary Fig. 4) with single stranded DNA (ssDNA) and dsRNA suggested that the sensing of the double helical structure of DNA was essential for the induction of R-BiP. Poly(dA:dT) dsDNA, an immunostimulatory reagent that mimics microbial dsDNA, strongly induced BiP arginylation (Fig. 2m–o) and nuclear localization of NFκB (Supplementary Fig. 5). Thus, BiP arginylation may be triggered when cells sense invading pathogenic dsDNA and activate innate immune responses.
Multiple Ca++-binding ER chaperones undergo both constitutive and DNA-induced arginylation at their N-termini
Bioinformatic analysis showed that many ER-targeted human proteins, including CRT, PDI, GRP94, and ERdJ5, cotranslationally and cotranslocationally acquire the potential to undergo N-terminal PTMs (Supplementary Tables 1–4). Their arginylation-permissiveness is evolutionarily conserved across various species. Consistently, earlier studies have shown that a portion of human CRT was present in the cytosol in an arginylated form45. To characterize N-terminal arginylation of these proteins, we raised antibodies to their arginylated forms, amongst which anti-R-PDI DH1 and anti-R-CRT DH1 antibodies to RDAPEEEDHVL and REPAVYFKEQ, respectively, showed sufficient specificity and avidity (Supplementary Fig. 2). In vivo arginylation assays showed that ATE11A7A mediates N-terminal arginylation of PDI and CRT at Nt-Asp18 and Nt-Glu18, respectively, which are exposed upon signal peptide cleavage (Fig. 2p). Unlike R-BiP, detectible amounts of R-PDI and R-CRT were constitutively generated in various cell lines (Fig. 2p), indicating their differential roles in the homeostasis of unstressed cells. Despite apparent differences among R-BiP, R-PDI, and R-CRT, their N-terminal arginylation was commonly triggered by cytosolic dsDNA (Fig. 2k, m, n, q) and proteasomal inhibition (see below), indicating a shared role in innate immune responses to invading microbes. These results suggest that the N-end rule pathway has a broad role in the turnover and functions of ER-residing proteins.
R-BiP is targeted to autophagosomes via p62 bodies
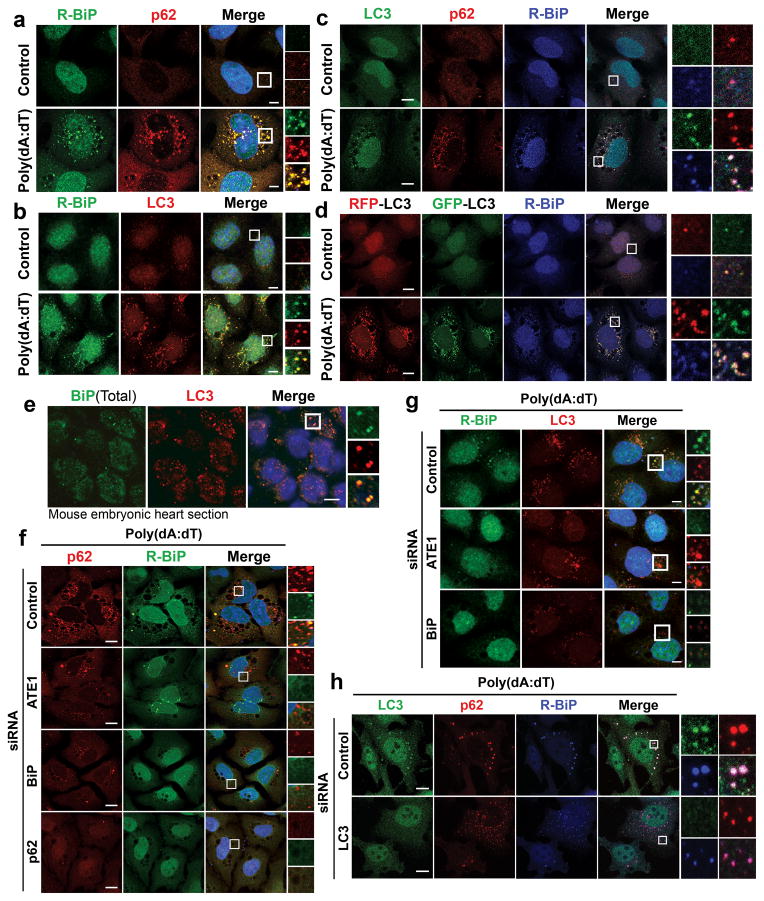
Immunoblotting analysis showed that DNA-induced arginylation of ER proteins correlated with the synthesis and activation of LC3 (Fig. 2k, o). Immunostaining showed that DNA-induced R-BiP formed cytosolic puncta with diameters of 0.1–1 μm that colocalized with puncta containing p62 (Fig. 3a) as well as LC3 (Fig. 3b). Colocalization of R-BiP puncta with p62 and LC3 puncta was confirmed in three-color costaining analysis (Fig. 3c) as well as in HeLa cells stably expressing RFP-GFP-LC3 (Fig. 3d and Supplementary Fig. 4). Within R-BiP+p62+ and R-BiP+LC3+ puncta, R-BiP puncta were smaller than and morphologically different from p62 and LC3 puncta, indicating that R-BiP is first targeted to p62 bodies and subsequently delivered to LC3-positive autophagosomes. Autophagic delivery of BiP was also observed on paraffin sections of mouse embryonic hearts (Fig. 3e). RNA interference assays showed that both ATE1 and BiP were required for optimal formation of p62 bodies (Fig. 3f) and LC3-positive autophagosomes (Fig. 3g), indicating the role of R-BiP in the induction of p62-mediated autophagy in response to poly(dA:dT). Reciprocally, p62-knockdown perturbed R-BiP delivery to autophagic vacuoles (Fig. 3f). By contrast, LC3-knockdown did not significantly affect the colocalization of R-BiP with p62 puncta (Fig. 3h). These results suggest that R-BiP is targeted to autophagosomes via p62 bodies and that N-terminal arginylation and R-BiP play a role in p62 delivery to autophagosomes.
Figure 3.
R-BiP is targeted to the autophagosome via p62 bodies. Scale bars, 10 μm. (a) Colocalization of cytoplasmic R-BiP puncta with p62 puncta in poly(dA:dT)-treated HeLa cells. (b) Colocalization of R-BiP puncta with LC3 puncta in HeLa cells stably expressing RFP-GFP-LC3 as determined by immunostaining of R-BiP in comparison with RFP signal (red) which represents LC3-positive autophagic vacuoles. (c) Three color colocalization analysis between R-BiP (blue), p62 (red), and LC3 (green) in poly(dA:dT)-treated HeLa cells. (d) HeLa cells expressing RFP-GFP-LC3 were subjected to three color colocalization analysis between R-BiP (blue), acid-resistant RFP (red), and acid-sensitive GFP (green). Most R-BiP puncta show a strong colocalization with LC3 puncta which are positive for both RFP and GFP, indicating the delivery of R-BiP to autophagosomes. (e) Immunohistochemistry of total BiP and LC3 on sections of mouse embryonic hearts at embryonic day 13.5, which reveals BiP puncta that colocalize with LC3 puncta. (f) RNA interference assay of ATE1, BiP, and p62 in poly(dA:dT)-treated HeLa cells, followed by colocalization analysis between R-BiP and p62. Note that knockdown of any of ATE1, BiP, and p62 disrupts the targeting of both R-BiP and p62 to autophagic vacuoles. (g) RNA interference assay of ATE1 and BiP in poly(dA:dT)-treated HeLa cells expressing RFP-GFP-LC3, followed by colocalization analysis between R-BiP and LC3. (h) RNA interference assay of LC3 in poly(dA:dT)-treated HeLa cells, followed by colocalization analysis between R-BiP, p62, and LC3. LC3-knockdown apparently did not affect significantly the delivery of R-BiP to p62 puncta.
Nt-Arg of R-BiP functions as a delivery determinant during R-BiP targeting to p62 and autophagosomes
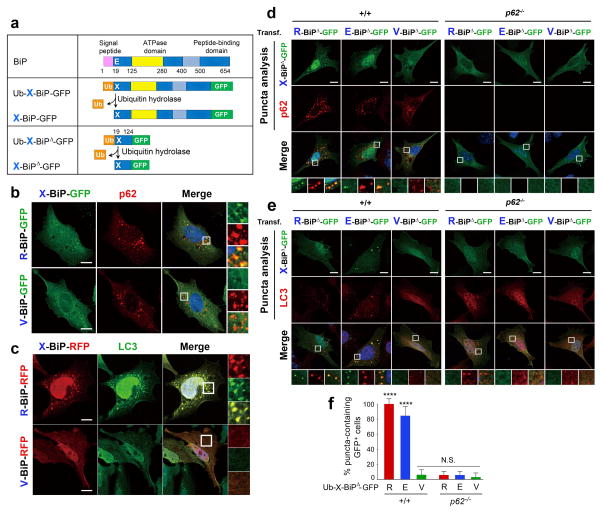
Little is known about the mechanism by which cargoes are selectively delivered to autophagy. Colocalization analyses showed that R-BiP-GFP, generated from Ub-R-BiP-GFP (Fig. 4a), formed cytosolic puncta that colocalize with p62 bodies (Fig. 4b) and LC3-positive autophagosomes (Fig. 4c). Glu19-to-Val mutation abolished BiP colocalization with autophagic components. To determine whether Nt-Arg is an autophagic delivery determinant, we removed the ATPase and substrate binding domains from X-BiP-GFP, leaving the first 106 residue fragment, Ub-X-BiP19–124-GFP (X= Glu, Arg, or Val) (Fig. 4a). R-BiP19–124-GFP (R-BiPΔ-GFP) and E-BiP19–124-GFP (E-BiPΔ-GFP) were readily targeted to p62 and LC3 puncta (Fig. 4d-f). Autophagic targeting of R-BiPΔ-GFP and E-BiPΔ-GFP was abolished in p62−/− MEFs (Fig. 4d-f), indicating that R-BiP delivery to autophagosomes requires p62. Moreover, Glu19-to-Val mutation abolished BiP colocalization with p62 and LC3 puncta (Fig. 4d-f). Thus, R-BiP Nt-Arg is a delivery determinant in p62-mediated macroautophagy.
Figure 4.
The Nt-Arg residue of R-Bip is a delivery determinant to the autophagosome. (a) A schematic diagram showing that the Ub fusion protein Ub-X-BiP-GFP is cotranslationally cleaved into Ub and X-BiP-GFP by Ub hydrolases. Also shown is how Ub-X-BiP19–124-GFP (Ub-X-BiPΔ-GFP) is cotranslationally cleaved into Ub and X-BiPΔ-GFP. (b) Colocalization analysis between X-BiP-GFP (X= Arg (an arginylated form of Glu-BiP, Arg-Glu-BiP) or Val (a Glu-to-Val mutant, Val-BiP-GFP)) and p62 in HeLa cells. X-BiP-GFP is produced in vivo from a precursor protein, Ub-X-BiP-GFP, which is elaborated in a. Scale bar, 10 μm. (c) An analogous colocalization assay with X-BiP-RFP and LC3. Scale bar, 10 μm. (d) Puncta colocalization analysis of X-BiP19–124-GFP (X-BiPΔ-GFP; X= Arg, Glu, and Val) and p62 in +/+ and p62−/− MEFs. Note that the ability of X-BiPΔ-GFP to form cytosolic puncta not only follows the N-end rule but also requires p62. Scale bar, 10 μm. (e) Puncta colocalization analysis of X-BiPΔ-GFP and LC3 in +/+ and p62−/− MEFs, which shows that the colocalization of R-BiP puncta with LC3 depends on both the N-end rule and p62. Scale bar, 10 μm. (f) Quantitation of d and e indicating that the ability of R-BiP to form cytosolic puncta depends on p62. The graph shows the percentage of BiPΔ-GFP-positive cells that form BiPΔ-GFP punta. Mean +/− s.d. of n=3 independent experiments in which 200 cells were analysed per experimental point. Statistical significance was calculated using a one-way ANOVA test (N.S. p > 0.05; **** p < 0.0001).
R-BiP binds p62
To determine whether R-BiP Nt-Arg binds p62, we performed X-peptide pulldown assays14 using synthetic X-BiP peptides (X= Arg-Glu (permanently arginylated), Glu (native), or Val (control)) (Fig. 5a). R-BiP peptide, but not E-BiP or V-BiP peptide, pulled down endogenous p62 from HEK293 cell extracts (Fig. 5b). To further demonstrate that Nt-Arg is a binding ligand to p62, we used 11-mer model N-end rule peptides, X-nsP4 (X= Arg, Phe, or Val), corresponding to N-terminal region of the Sindbis virus polymerase nsP410. P62 preferentially bound Arg-nsP4 compared with Phe-nsP4 and Val-nsP4 (Fig. 5c). Thus, R-BiP binds p62 through the N-end rule interaction of its Nt-Arg.
Figure 5.
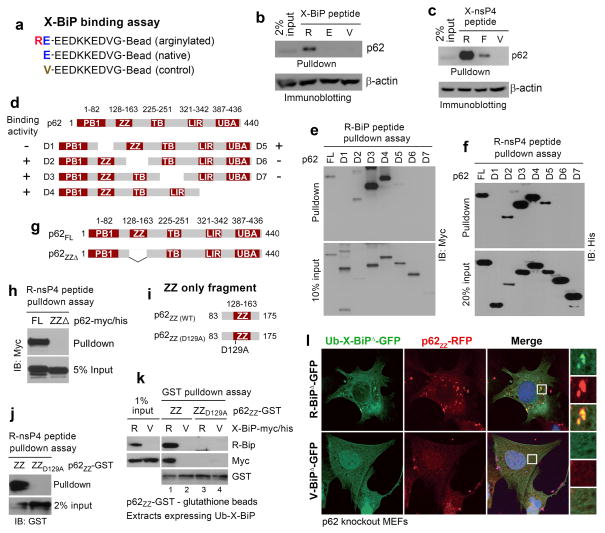
The Nt-Arg residue of R-BiP binds to the ZZ domain of p62. (a) The sequences of X-BiP peptides used to pulldown p62. (b) The Nt-Arg residue of R-Bip binds to p62. X-BiP peptides crosslinked to beads were used to pulldown p62 from HEK293 cell extracts transiently expressing p62. (c) Similar to b except that X-nsP4 peptides (X= Arg, Phe, and Val) were used. (d) A diagram showing C-terminally (D1-D4) and N-terminally (D5-D7) deleted p62 mutant proteins. (e) R-BiP peptide pulldown assay with serially deleted p62 mutants. (f) R-nsP4 peptide pulldown assay with serially deleted p62 mutants. (g) A diagram showing wild-type p62 and a ZZ-deletion mutant (p62ZZΔ). (h) R-nsP4 pulldown assay with wild-type p62 and p62ZZΔ. (i) A diagram showing a ZZ-only fragment (p62ZZ(WT), #83–175) and its mutant (p62ZZ(D129A)) in which the conserved Asp129 was mutated to Ala. (j) R-nsP4 peptide pulldown assay using p62ZZ(WT) and p62ZZ(D129A). (k) A GST-pulldown assay combined with X-BiP pulldown assay. A p62-GST fusion, p62ZZ-GST or p62ZZ(D129A)-GST, immobilized on gluthathione beads was incubated with HEK293 extracts expressing Ub-X-BiP-myc/his (X= Arg or Val), followed by centrifugation and immunoblotting analysis of X-BiP-myc/his that bound to p62 ZZ fragments. Note that R-BiP-myc/his binds to wild-type ZZ-only fragment but not to D129A-ZZ fragment. No binding was detected with V-BiP-myc/his. (l) Colocalization assay of X-BiPΔ-GFP (X= Arg or Val) and p62ZZ-RFP coexpressed in p62−/− MEFs. X-BiPΔ-GFP was cotranslationally generated from Ub-X-BiPΔ-GFP using the Ub fusion technique. Note that R-BiPΔ-GFP, but not V-BiPΔ-GFP, forms cytosolic puncta colocalizing with p62ZZ-RFP.
The ZZ motif of p62 is the ligand-binding domain for Nt-Arg
To identify the domain of p62 that binds BiP Nt-Arg, we constructed C-terminally (D1-D4) or N-terminally (D5-D7) deleted p62 mutants (Fig. 5d). Both R-BiP (Fig. 5e) and R-nsP4 (Fig. 5f) peptides pulled down D2, D3, D4 and D5 but not D1, D6, and D7, suggesting that Nt-Arg binds a region spanning ZZ domain, a poorly characterized zinc finger motif35. Consistently, a p62 mutant lacking ZZ domain (residues 128–163) no longer bound R-nsP4 peptide (Fig. 5g, h). GST-pulldown assays (Fig. 5i–k) showed that a 93-residue ZZ-only fragment (residues #83–175), p62ZZ-GST (Fig. 5i), was sufficient to bind R-BiP (Fig. 5k, lane 1). The interaction between p62ZZ-GST and BiP was abolished by Glu19-to-Val mutation (Fig. 5k, lanes 2 vs. 1). Similarly, the alanine (Ala) mutation of the conserved Asp129 residue within ZZ domain abolished such a binding activity of p62ZZ (Fig. 5k, lanes 3 vs. 1). Peptide pulldown assays independently demonstrated that R-nsP4 peptide also binds p62ZZ-GST but not its D129A mutant (Fig. 5j). Immunostaining analysis of p62−/− MEF cells showed that a 93-residue ZZ-only fragment fused with RFP, p62ZZ-RFP (#83–175), formed puncta colocalizing with R-BiPΔ-GFP but not V-BiPΔ-GFP (Fig. 5l). Thus, p62 ZZ motif is the ligand binding domain for Nt-Arg.
Nt-Arg induces p62 aggregation
Little is known about the mechanism by which p62 aggregation is regulated during autophagic induction. To determine the biological consequences of Nt-Arg binding to p62, we added the dipeptide Arg-Ala (type-1) in comparison with other dipeptides bearing N-terminal His, Lys (type-1), Phe, Trp, Try (type-2), or Ala (stabilizing) to HEK293 cell extracts expressing full-length p62 and monitored the conversion of p62 into oligomers and aggregates using nonreducing SDS-PAGE. When compared with other N-end rule dipeptides, we found that Arg-Ala strongly and selectively induced the oligomerization and aggregation of p62 (Fig. 6a-c). Ala-Arg, a control dipeptide bearing Arg at the second position, did not show such activity. These results suggest that Nt-Arg binding to p62 induces self-oligomerization and aggregation of p62 in vitro, consistent with our data that ATE1 is required for optimal p62 puncta formation in response to poly(dA:dT) (Fig. 3f).
Figure 6.
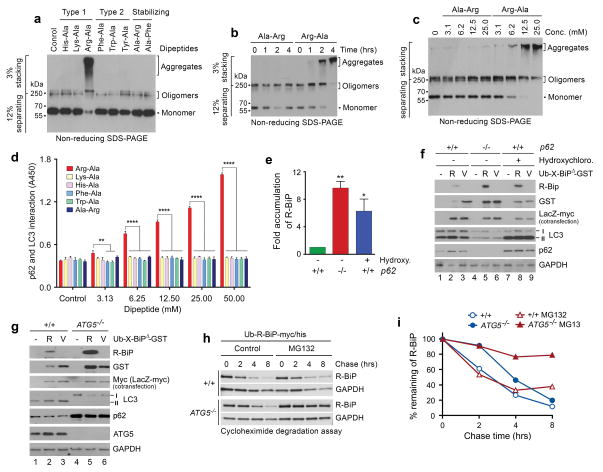
The Nt-Arg residue induces oligomerization and aggregation of p62 in vitro. (a) In vitro oligomerization/aggregation assays of HEK293 cell lysates expressing p62-myc/his. The dipeptide Arg-Ala (type-1) at a final concentration of 20 mM was added to the extracts (1 μg) in comparison with other dipeptides. (b, c) Similar to a. Arg-Ala was compared with Ala-Arg in a time-dependent (b) and dose-dependent (c) manner. (d) The p62-LC3 interaction assay shows that Nt-Arg of Arg-Ala promotes the interaction of p62 with LC3-GST. LC3-GST (3 μg) was immobilized to gluthathione (GSH)-coated wells and incubated with HEK293 cell extracts (20 μg) expressing p62. Following incubation with dipeptides for 2 hrs, bound p62 was detected using anti-p62 antibody. Quantification is of n = 3 independent experiments; bars represent mean ± s.d. Statistical significance was calculated using a one-way ANOVA test (N.S. P > 0.05; ** P < 0.01; **** P < 0.0001. (e) Quantitation of f. Quantification is of n = 3 independent experiments; bars represent mean ± SEM. Statistical significance was calculated using a one-way ANOVA test (* P < 0.05; ** P < 0.01). (f) R-BiPΔ-GST, but not V-BiPΔ-GST, is metabolically stabilized by p62 knockout or pharmaceutical inhibition of autophagy. Shown is immunoblotting of X-BiPΔ-GST (X= Arg or Val) in +/+ and p62−/− MEFs with or without the treatment of 25 μM hydroxychloroquine for 16 hrs. Note that the level of R-BiPΔ-GST (as determined by GST immunoblotting) is lower as compared with V-BiPΔ-GST because of its degradation (lanes 2 vs. 3) and increased in p62-deficient cells (lanes 2 vs. 5) or by the treatment of hydroxychloroquine. (g) R-BiPΔ-GST, but not V-BiPΔ-GST, is metabolically stabilized in ATG5−/− MEFs. Shown is immunoblotting of X-BiP19–124-GST (X-BiPΔ-GST, X= Arg or Val) in +/+ and ATG5−/− MEFs. Note that the level of R-BiPΔ-GST, as determined by GST immunoblotting, is lower than V-BiP19–124-GST (lanes 2 vs. 3) and increased in ATG5-deficient cells (lanes 2 vs. 5). (h) Cycloheximide degradation assay of R-BiP-myc/his generated from Ub-R-BiP-myc/his in +/+ and ATG5−/− MEFs in the absence or presence of 5 μM MG132. Cells were treated with 50 μg/ml cycloheximide, followed by time-course immunoblotting of R-BiP. (i) Quantitation of h. Shown is a representative of two independent experiments.
Nt-Arg increases p62 interaction with LC3
The mechanism by which the interaction of cargo-associated p62 with LC3 on autophagic membranes is regulated remains unclear35. To determine whether Nt-Arg binding to p62 increases p62 interaction with LC3, we performed GST pulldown-coupled ELISA wherein HEK293 cell extracts ectopically expressing p62 were incubated with LC3-GST immobilized on the surface of glutathione-coated wells in the presence of the aforementioned dipeptides (Fig. 6d). Compared with other dipeptides, only Arg-Ala markedly increased p62 interaction with LC3 (Fig. 6d). Our in vitro (Figs. 5 and 6) and in vivo (Figs. 3 and 4) assays collectively suggest that Nt-Arg binding to p62 induces an allosteric conformational change, exposing PB1 and LIR domains, which, in turn, promotes self-oligomerization and LC3 interaction, respectively.
Nt-Arg of R-BiP is an autophagic degron
Little is known about PTM-generated degrons that mediate selective proteolysis by autophagy. To determine whether R-BiP is a selective substrate of macroautophagy, we monitored the decay of X-BiPΔ-GST generated from Ub-X-BiPΔ-GST (Fig. 6e–g). R-BiPΔ-GST exhibited a shorter half-life than V-BiPΔ-GST as determined by the levels of GST (Fig. 6f, lanes 2 vs. 3). As quantified in Figure 6e, the degradation of R-BiPΔ-GST was inhibited by p62-knockout (Fig. 6f, lanes 5 vs. 2) or the pharmaceutical blockage of autophagy (lanes 8 vs. 2). Autophagic degradation of R-BiPΔ-GST was further demonstrated by its accumulation in ATG5−/− MEFs (Fig. 6g, lanes 5 vs. 2). Finally, cycloheximide degradation assays confirmed that R-BiP-myc/his was short-lived in MEFs and metabolically stabilized by ATG5-knockout alone and, synergistically, when combined with proteasomal inhibition (Fig. 6h, i). These results suggest that R-Bip is selectively degraded by autophagy through the activity of Nt-Arg as a cis-acting autophagic degron.
N-terminal arginylation of ER proteins is signaled by and associated with cytosolic misfolded proteins destined to autophagy
We tested whether N-terminal arginylation of ER proteins is induced by Ub conjugates destined to autophagy. Immunoblotting analysis showed that poly(dA:dT) coinduced ubiquitination and R-BiP (Fig. 7a). Immunostaining analysis showed that poly(dA:dT)-induced Ub-conjugates formed cytosolic puncta that colocalize with R-BiP as well as p62 (Fig. 7b). Consistently, a screening with various stressors identified prolonged proteasomal inhibition as a stress type that strongly induces R-BiP (Fig. 7c). The induction of R-BiP, R-CRT, and R-PDI under proteasomal inhibition became synergistic when cotreated with thapsigargin (Fig. 7d, e), suggesting that ER stress accelerates the supply of the precursor, lumenal BiP, for N-terminal arginylation. In addition to proteasomal inhibition and ER stress, geldanamycin, an Hsp90 inhibitor that facilitates the formation of misfolded proteins, induced R-BiP (Fig. 7f). These results suggest that N-terminal arginylation of ER proteins is signaled by cytosolic misfolded proteins which are tagged with Ub but redirected to autophagy.
Figure 7.
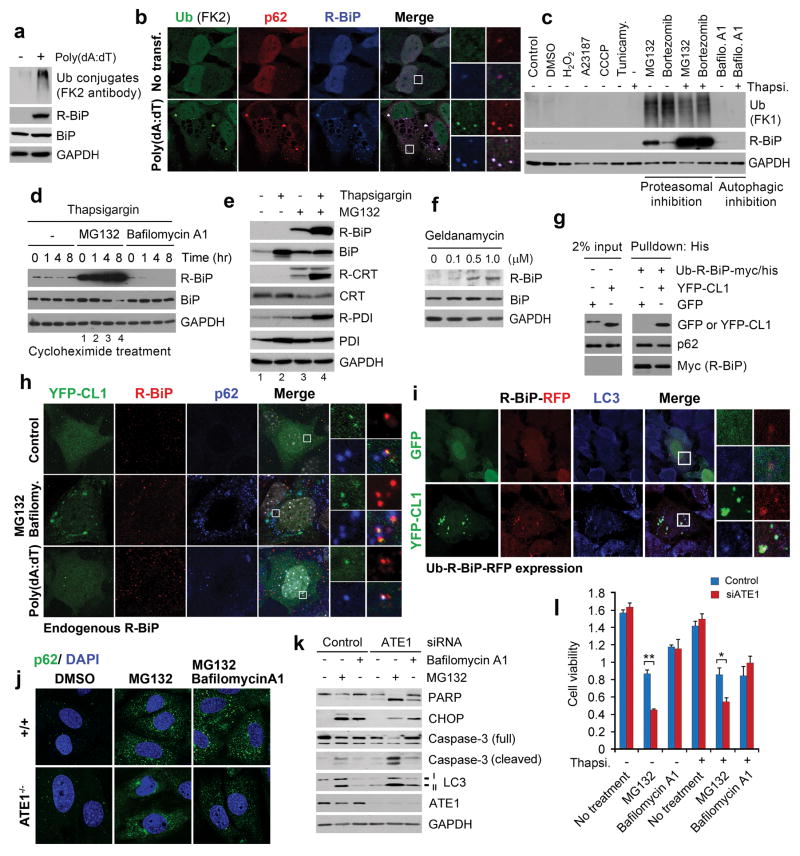
R-BiP is induced by and associated with cytosolic misfolded proteins, and ATE1-deficient cells are hypersensitive to misregulation of protein quality control. (a) Immunoblotting analysis of R-BiP and Ub conjugates in HeLa cells treated with 0.4 μg/ml poly(dA:dT) for 16 hrs. (b) Colocalization assay with antibodies specific to Ub conjugates (FK2 antibody), p62, and R-BiP in HeLa cells treated with 1 μg/ml poly(dA:dT) for 21 hrs. Scale bar, 10 μm. (c) HeLa cells were treated with various stressors for 24 hrs as described in Methods and subjected to immunoblotting analysis of R-BiP and Ub conjugates using FK1 antibody. (d) HeLa cells were treated with 10 μM MG132 in combination with 200 nM thapsigargin or 200 nM bafilomycin A1 for 16 hrs. The cells were treated with 10 μg/ml cycloheximide, followed by immunoblotting assay. (e) Arginylation of ER proteins is synergistically induced by proteasomal inhibition and ER stress. HeLa cells were treated with 10 μM MG132 and 100 nM thapsigargin for 18 hrs. (f) The treatment of geldanamycin, an inhibitor of the HSP90, for 17 hrs results in co-induction of autophagy with arginylation of ER-residing chaperons. (g) Measurement of the interaction between R-BiP and CL1-YFP, a model substrate that undergoes spontaneous misfolding. See Methods for experimental details. (h) Colocalization assay of YFP-CL1 with R-BiP and p62. MEFs ectopically expressing YFP-CL1 was treated with 1 μg/ml poly(dA:dT) alone for 18 hrs or 10 μM MG132 and 200 nM bafilomycin A1 for 6 hrs, followed by immunostaining of endogenous R-BiP and p62 in comparison with YFP-CL1 fluorescence. Scale bar, 5 μm. (i) Ub-R-BiP was coexpressed with YFP-CL1 or GFP in HeLa cells, followed by fluorescence analysis. (j) Puncta formation assay of p62 in +/+ and ATE1−/− MEFs treated with 10 μM MG132 and/or 0.2 μM bafilomycin A1 for 6 hrs. (k) Control and ATE1-knockdown cells were treated with 10 μM MG132 and/or 0.2 μM bafilomycin A1 for 18 hrs, followed by immunoblotting analysis. (l) MTT assay of control and ATE1-knockdown cells treated with various stressors, including 10 μM MG132. Mean +/− s.d. of n=3 independent experiments in which 10,000 cells in a 24-well plate were analysed per experimental point. Statistical significance was calculated using a two-way ANOVA test (* p < 0.05; ** p < 0.01).
Given the intrinsic ability of BiP to bind and deliver terminally misfolded proteins to ER-associated protein degradation (ERAD)37, we determined whether R-BiP is associated with YFP-CL146, a model cytosolic substrate of spontaneous misfolding (Fig. 7g-i). In MEFs treated with MG132 and bafilomycin A1, YFP-CL1 formed cytosolic puncta that colocalize with R-BiP as well as p62 (Fig. 7h). A similar colocalization of YFP-CL1 and R-BiP with p62 and LC3 was observed in MEFs treated with poly(dA:dT) (Fig. 7h) or expressing Ub-R-BiP (Fig. 7i). Finally, the molecular interaction between R-BiP and YFP-CL1 was confirmed by using coprecipitation assays (Fig. 7g). These results suggest that the cargoes of R-BiP, an ER-derived counterpart of cytosolic Hsp70, include cytosolic misfolded proteins destined to autophagy.
Arginylation-deficient cells are hypersensitive to the misregulation of protein quality control
To determine the physiological importance of Nt-Arg in stress responses to misregulation of protein quality control, we treated +/+ and ATE1−/− MEFs with MG132 for 6 hrs and monitored the formation of p62 puncta. Consistent with the finding that Nt-Arg is an autophagic delivery determinant (Fig. 4) as well as an activation ligand to p62 (Figs. 5 and 6), immunostaining analysis showed that ATE1−/− MEFs were impaired in the formation of p62 puncta under proteasomal inhibition (Fig. 7j), suggesting that Nt-Arg plays an important role in p62-dependent autophagic delivery of cytosolic misfolded proteins. In addition, ATE1-knockdown rendered MEFs hypersensitive to prolonged proteasomal inhibition (Fig. 7l) resulting in apoptotic death through the cleavage of caspase 3 and PARP (Fig. 7k). These results indicate that N-terminal arginylation by ATE1 is essential for coping with cellular stresses caused by excessive misfolded proteins.
DISCUSSION
Although many ER-residing proteins have been found in various cellular compartments41,42, 47,48, the mechanisms underlying their functions remain poorly. Here, we show that a large number of ER-targeted proteins acquire the potential to undergo N-terminal arginylation and other N-end rule PTMs upon signal peptide cleavage. These include major Ca++-binding folding factors whose arginylation-permissiveness is conserved throughout evolution. By raising arginylation-specific antibodies, we show that BiP, PDI, and CRT are the physiological substrates of ATE1-encoded R-transferase, suggesting that the N-end rule pathway may have a broad role in the regulation of ER proteins. One fundamental question not addressed in this study is whether these arginylation substrates are released into the cytosol through a translocon-like retrotranslocation channel47,48 and/or as endosome-like vesicles.
In part because of their low abundance in unstressed cells, it has been challenging to characterize the metabolic fates of ER-derived proteins in non-ER compartments. We now demonstrate that cytosolic R-BiP is selectively delivered to autophagic vacuoles via p62 bodies (Figs. 3–4). The autophagic targeting requires p62 (Figs. 3f; 4d, e) and involves the binding of R-BiP Nt-Arg to p62 (Fig. 5a–c). The autophagic/signaling adaptor p62 is known to exert its functions through multiple domains such as PB1 (for self-oligomerization), ZZ, TB (TRAF6-interacting), LIR (LC3-interacting), and UBA (Ub-interacting) 34. Although these domains have been extensively studied in signaling and autophagic processes34, the functions of ZZ, a C2H2 zinc finger domain, has remained murky. Our X-peptide and GST binding assays (Fig. 5d–k) and colocalization assays (Fig. 5l) now show that ZZ is the ligand-binding domain for Nt-Arg, making p62 an N-recognin in autophagy. Moreover, our aggregation and binding analyses (Fig. 6) suggest that Nt-Arg binding to p62 induces self-oligomerization and aggregation and increases p62 interaction with LC3. These biochemical consequences are consistent with our data that R-BiP is delivered to autophagosomes and degraded by lysosomal hydrolases (Fig. 6e–i). Thus, Nt-Arg, a well characterized degron in the UPS1–5, is an autophagic degron as well. One intriguing observation with R-BiP turnover was that whereas a short N-terminal fragment of R-BiP was readily degraded by autophagy (Fig. 6e–g), endogenous R-BiP exhibited relatively longer half-life (Fig. 2i). The apparent difference could be because full-length R-BiP has multiple domains that interact with other molecules, resulting in cytosolic retention prior to targeting for lysosomal degradation. In summary, we propose a model (Fig. 8) that the binding of R-BiP Nt-Arg to the ZZ domain of p62 “activates” p62 through an allosteric conformational change, which exposes PB1 domain that mediates self-oligomerization and LIR domain that mediates the interaction with LC3 on autophagic membranes. In this autophagic regulatory mechanism, Nt-Arg is a delivery determinant to autophagic vacuoles, an activating ligand to p62, and a trans-acting autophagic degron to BiP and other arginylated ER proteins. Given the emerging role of Nt-Arg in autophagic degradation, another important question concerns the identity of cargoes of arginylated ER proteins. We demonstrate that one signal that coinduces N-terminal arginylation of ER proteins together with ubiquitination and autophagy is cytosolic misfolded proteins destined to p62-dependent macroautophagy, which are generated during cellular stress responses such as innate immune responses to cytosolic foreign dsDNA (Fig. 7a–f). Consistently, we find that R-BiP associates with cytosolic misfolded proteins in vitro and in autophagic vacuoles (Fig. 7g–i), indicating the role for R-BiP in autophagy-mediated homeostasis of proteins and nonproteinaceous cargoes.
Figure 8.
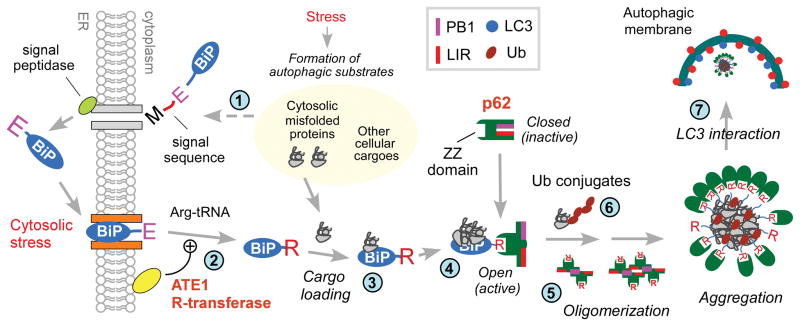
A model illustrating the role of the N-end rule pathway in N-terminal arginylation of ER-residing proteins and the ligand-mediated regulation of autophagy in stressed cells. In this model, ER residents and clients acquire cotranslationally and cotranslocationally the degrons of the N-end rule pathway, which normally remain separated from cytosolic N-end rule machinery. N-terminal arginylation of ER-residing proteins, especially BiP, is induced by various stress signals such as cytosolic misfolded proteins that are initially tagged with Ub but cannot be readily processed by the proteasome (Step 1). One physiological stress signal that induces N-terminal arginylation of ER proteins is the presence of cytosolic foreign dsDNA which triggers innate immune responses. N-terminal arginylation is thought to occur when the N-terminal residue of the substrates is exposed to the cytosolic surface of the ER membrane (Step 2), which may facilitate the cytosolic relocalization of arginylated ER proteins. We do not exclude the possibility that N-terminal arginylation occurs after the substrates complete their cytosolic relocalization. Arginylated ER proteins relocated to non-ER compartments appear to have both shared and distinct functions and metabolic fates. At least a subpopulation of cytosolic R-BiP appears to be associated with its cargoes such as cytosolic misfolded proteins (Step 3). R-BiP, alone or loaded with its cargo, binds to the ZZ domain of p62 through the N-end rule interaction of its Nt-Arg (Step 4). This induces an allosteric conformational change of p62, exposing PB1 and LIR domains. PB1 domain promotes self-oligomerization and aggregation of p62 (Step 5), together with cargoes such as cytosolic misfolded proteins (Step 6). LIR domain mediates the interaction of p62 with LC3 on the autophagic membranes (Step 7). In this model, the Nt-Arg residue acts as an autophagic delivery determinant to autophagosomes for BiP and its cargoes, an activating ligand to p62, and an autophagic degron for BiP and the cargoes of R-BiP and p62.
SUPPLEMENTARY METHODS
Plasmids and other reagents
The plasmids encoding ATE1 R-transferase isoforms are described50. To construct a plasmid encoding full-length BiP-flag-KDEL, in which the C-terminal KDEL ER retention signal was replaced with flag-KDEL, the BiP-flag-KDEL fragment was amplified from pENTR/hBiPc (Invitrogen) using polymerase chain reaction (PCR) and inserted into the vector pcDNA3.0 at the HindIII/BamHI-KDEL site. A Glu19-to-Val point mutation in BiP was generated using the QuickChange XL method (Stratagene). The plasmids encoding Ub-X-BiP-GFP and Ub-X-BiP19–124-GFP were constructed into pEGFP-N3 at the BamHI/HindIII site using PCR amplification. To construct cDNA encoding full-length BiP-myc/his, the BiP fragment was amplified from pENTR/hBiPc (Invitrogen) using PCR and inserted into the pcDNA3.1-myc/his vector at the EcoRV-XhoI site. The plasmids encoding Ub-X-BiP-myc/his were constructed into pcDNA3.1-myc/his at the BamHI/ XbaI site using PCR amplification. To construct a plasmid encoding full-length human p62 tagged with myc and his, a 1,320-bp cDNA fragment was PCR-amplified from the hMU012675 clone (21C Frontier Human Gene Bank), digested with restriction enzymes EcoRI and XhoI, and subcloned into pcDNA.3.1/myc-his. To construct a series of deletion mutants (D1-D7, CD1-CD9, and ND1-ND6) of human p62, we amplified appropriate DNA fragments from the hMU012675 clone and subcloned them into pcDNA 3.1/myc-His through a similar strategy. These constructed plasmids were confirmed by sequencing.
The following plasmids are commercially available: N3-EGFP (Addgene, cat #6080-1), Ub-HA (Addgene, 17608: pRK5-HA-ubiquitin-WT), Ub-mt-HA (Addgene, 17603: pRK5-HA-ubiquitin-KO), YFP-CL1 (Addgene, 11950), IgK-IFN β-Luc (Addgene, 14886), and lenillar-luc (Promega, E2231, pRL-SV40 vector). The NHK-GFP plasmid was a gift from Dr. Nobuko Hosokawa (Kyoto University, Japan). Other reagents used in this study were purchased as follows: thapsigargin (Sigma, T9033), digitonin (Sigma, D141), bafilomycin A1 (Sigma, B1793), geldanamycin (Sigma, G3381), MG132 (Calbiochem, 474790), poly(dA:dT) (Invivogen, tlrl-patn), 5′ppp-RNA (Invivogen, tlrl-3prna), salmon sperm DNA (Sigma, D1626), RNAiMax (Invitrogen, 13778), Lipofectamine 2000 (Invitrogen, 11668), Lipofectamine LTX (Invitrogen, 15338), protein A/G plus agarose bead (Santa Cruz, sc2003), carbonyl cyanide chlorophenylhydrazone (Sigma, C2759), A23187 (Sigma, C7522), bortezomib (BTZ, Selleckchem, S1013), hydrogen peroxide (Sigma, H1009), Alexa Fluor 594 microscale protein labeling kit (Life Technologies, A30008), and Dual-luciferase reporter assay system (Promega, E1910).
Antibodies
The antibodies used in this study are as follows: rabbit monoclonal anti-BiP (Cell Signaling, cat. #3177, 1:1,000), rabbit polyclonal anti-GFP (Abcam, ab290, 1:3,000), mouse monoclonal anti-LC3 (Santa Cruz, sc271625, 1:1,000), rabbit polyclonal anti-GAPDH (Santa Cruz, sc25778, 1:2,000), mouse monoclonal anti-flag (Sigma, f1804, 1:3,000), mouse monoclonal anti-ATE1 (Santa Cruz, sc-271220, 1:500), rabbit monoclonal anti-IRF-3 (Cell Signaling, 11904P, 1:500), rabbit monoclonal anti-phospho-IRF-3 (Ser396) (Cell Signaling, 4947S, 1:500), rabbit polyclonal anti-NFkB p50 (Santa Cruz, SC-7178, 1:50), mouse monoclonal anti-KDEL (Abcam, ab12223, 1:200), mouse monoclonal anti-calreticulin/CRT (BD Biosciences, 612136, 1:5,000), mouse monoclonal anti-polyubiquitylated FK2 clone (Millipore, cat. #04-263), and rabbit polyclonal anti-PDI (Cell Signaling, 2446S, 1:1,000), mouse monoclonal anti-p62 (Santa Cruz, SC-28359). The following secondary antibodies were used: alexa fluor 488 goat anti-rabbit IgG (Invitrogen, A11034, 1:200), alexa fluor 488 goat anti-mouse IgG (Invitrogen, A11029, 1:200), alexa fluor 555 goat anti-rabbit IgG (Invitrogen, A21429, 1:200), alexa fluor 555 goat anti-mouse IgG (Invitrogen, A21424, 1:200), alexa fluor 633 goat anti-rabbit IgG (Invitrogen, A21071, 1:200), alexa fluor 633 goat anti-mouse IgG (Invitrogen, A21052, 1:200), anti-rabbit IgG-HRP (Cell Signaling, 7074, 1:10,000), and anti-mouse IgG-HRP (Cell Signaling, 7076, 1:10,000).
Antibodies to the arginylated species of BiP, CRT, and PDI
Rabbit polyclonal antibodies (anti-R-BiP DH1, anti-R-CRT DH1, and anti-PDI DH1) specific for the arginylated forms of BiP, CRT, and PDI were respectively raised using the peptide sequences REEEDKKEDVGC, REPAVYFKEQ, and RDAPEEEDHVL, corresponding to the N-terminal sequences of the arginylated mature proteins through a custom service at AbFrontier Inc. (Seoul, South Korea). The immunizations with these peptides were performed in rabbit and boosted with incomplete Freund’s adjuvant (IFA) at 3-week intervals. Following IgG purification using immobilized Protein A, the antibodies were subjected to two-step affinity chromatography. First, the antibodies were negatively purified using an unarginylated peptide, EEEDKKEDVGC, EPAVYFKEQ, or DAPEEEDHVL, to remove nonspecific IgG. The antibodies in the flow-through from the first affinity chromatography were positively purified using an arginylated peptide, REEEDKKEDVGC, REPAVYFKEQ, or RDAPEEEDHVL, by eluting in 0.1 M citric acid. The specificity of the purified antibodies was validated using immunoblotting analysis of the cells cultured in the presence or absence of arginylation-inducing stressors. The specificity was independently characterized using a peptide binding assay (see Supplemental Methods). Specifically, each of the peptides (unmodified and arginylated) for BiP, CRT, and PDI was diluted to 2 μg/ml in coating buffer (0.1 M carbonate pH 9.6) and immobilized on a 96-well plate by incubating at 4°C for overnight, followed by incubation in blocking buffer A (2% skim milk in TBS-T) at 37°C for 1 hr. After washing with TBS-T, the immobilized peptide was incubated at 37°C for 2 hrs in blocking buffer containing a primary antibody to BiP, CRT, or PDI in the presence or absence of a competing peptide, followed by incubation with a goat anti-rabbit secondary antibody conjugated to horseradish peroxide. The binding of antibody to the immobilized peptide was measured in optical density (O.D.) at 450 nm.
Subcellular fractionation
To obtain cytosolic proteins, cells were trypsinized and pelleted by centrifugation at 1,500 X g. The plasma membranes of the harvested cells were permeabilized using 0.01% digitonin, a glycoside from Digitalis purpurea, in lysis buffer (110 mM KOAc, 25 mM K-HEPES, pH7.2, 2.5 mM Mg (OAc)2, and 1 mM EGTA). Digitonin-permeabilized cells containing the bulk ER were pelleted by centrifugation for 5 min at 1,000 X g. Following centrifugation, the resulting supernatant was centrifuged at 15,000 X g to obtain a cytosolic fraction that contains soluble cytosolic proteins and some autophagic vacuoles that have been released from the digitonin-permeabilized cells. As an alternative way to obtain cytosolic proteins, cells were swollen in hypotonic lysis buffer (10 mM KOAc, 10 mM KHEPES, pH7.2, 1.5 mM Mg(OAc)2), and their plasma membranes were torn off using a Dounce homogenizer. After adjusting the buffer to 0.25 M sucrose, the cell suspension was centrifuged at 1,000 X g for 10 min to discard unbroken cells and nuclei, followed by another centrifugation at 12,000 X g for 15 min to remove mitochondria. The resulting supernatant was ultracentrifuged at 100,000 X g for 1 hr to separate soluble cytosolic proteins from microsomes that represent vesicular structures containing pieces of the ER.
Cell culture, immunoblotting, and immunoprecipitation
HeLa, PC3, and HEK293 cells as well as +/+ and ATE1−/− MEFs were cultured in DMEM (GIBCO) supplemented with 10% FBS (GIBCO) in a 5% CO2 incubator. For immunoblotting, cells were lysed using NETN lysis buffer (100 mM NaCl, 20 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, and 0.5 % Nonidet P-40) or RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS). Whole cell lysates were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes. For immunoprecipitation, cell lysates in NETN lysis buffer were incubated with primary antibody at 4°C for 4 hrs and subsequently with protein A/G agarose beads (Santa Cruz). The proteins bound to antibody-bead complexes were washed with cold NETN buffer and separated by SDS-PAGE for immunoblotting analysis.
To identify chemical stressors that induce R-BiP, HeLa cells were treated with the following reagents for 24 hrs: 500 μM H2O2; 2 μg/ml A23187, calcium ionophore; 10 μM CCCP (carbonyl cyanide m-chlorophenyl hydrazone), a chemical inhibitor of oxidative phosphorylation; 100 nM thapsigargin; 5 μM MG132; 5 μM bortezomib (PS-341/Velcade), a proteasome inhibitor; and 0.1 μM bafilomycin A1, an autophagic inhibitor that inhibits vacuolar H+ ATPase (V-ATPase).
To determine the interaction of R-BiP with YFP-CL1, a model substrate of spontaneous misfolding, HeLa cells exogenously expressing Ub-R-BiP-myc/his were lysed in a native purification buffer (50 mM NaH2PO4, pH8.0, 250 mM NaCl and 10 mM imidazole with a cocktail of protease and phosphatase inhibitors (Roche)) through two cycles of freezing in liquid nitrogen and thawing in a 42°C water bath. Genomic DNA was sheared by passing the extracts through a 26-gauge needle four times. The resulting lysates were centrifuged at 3,000 × g for 15 min to pellet the cellular debris. Total 1 mg proteins were incubated with 50 μl of 50% Ni-NTA agarose beads for 1 hr with gentle agitation, and then the beads were washed with wash buffer (50 mM NaH2PO4, pH8.0, 250 mM NaCl and 20 mM imidazole (Roche)) for 10 min at 4°C with gentle agitation; washing was repeated three times. The beads were then incubated with HeLa cell extracts (500 μg protein) containing either exogenously expressed GFP or YFP-CL1 in binding buffer (20 mM Tris-HCl, pH 7.6, 125 mM NaCl, 15 mM imidazole and 1% Nonidet P-40 with a cocktail of protease and phosphatase inhibitors (Roche)) for 3 hrs with gentle agitation. The beads were washed with the binding buffer for 5 min at 4°C with gentle agitation; washing was repeated three times. The proteins bound to His-NTA agarose beads were dissociated in 2X SDS sample buffer, heated at 100 °C for 5 min and separated on a SDS-PAGE.
Protein degradation assays
For pulse chase analysis, cells at 80% confluence were treated with 100 nM thapsigargin for 6 hrs to induce the transcription of BiP and pulse-labeled with 35S-methionine/cysteine (35S-Express protein labeling mix; Perkin-Elmer) for 12 min, followed by a chase in the presence of cycloheximide, preparation of extracts, immunoprecipitation, NuPAGE 10%-Bis-Tris SDS-PAGE (Invitrogen), autoradiography, and quantitation of 35S (Molecular Imager FX system, Bio-Rad, Hercules, CA). For cycloheximide-chase degradation assay, cells at 80% confluence were treated with 10 μg/ml cycloheximide. At indicated time points, cells were lysed on ice for 30 min in RIPA buffer containing a protease inhibitor cocktail, followed by centrifugation for 20 min at 15,000 g. After centrifugation, 10 μg of total protein was subjected to immunoblotting.
RNA interference analysis
Cells in a 6-well plate (1 × 106/well) were transfected with 100 pmol of siRNA using the RNAi Max reagent (Invitrogen). Pre-designed Silencer Select siRNAs (Invitrogen) were used to knockdown ATE1, BiP, and p62 using the following sequences: siATE1 #204 (HSS117-204-sense, ACCCACCAUCUUUGUUUCCACCAAA; HSS117-204-antisense, UUUGGUGGAAACAAAGAUGGUGGGU), siGRP78/BiP (sense, AGUGUUGGAAGAUUCUGAUTT; antisense, AUCAGAAUCUUCCAACACUTT), and siSQSTM1 (HSS113117-sense, AUAGUUCUUGGUCUGCAGGAGCCUG; HSS113117-antisense, CAGGCUCCUGCAGACCAAGAACUAU).
Immunocytochemistry
Immunocytochemistry of an arginylated form of ER-residing proteins in cultured cells was performed using a standard procedure51. Briefly, the cells were washed with PBS twice, fixed in 4% paraformaldehyde (PFA) at room temperature for 10 min, treated with ice-cold methanol for 2.5 min, and incubated in blocking solution (5% BSA in PBS), followed by incubation with primary antibody and subsequently Alexa Fluor -labeled goat anti-rabbit/mouse IgG (Invitrogen). BiP is stained using goat anti-rabbit IgG conjugated with an Alexa Fluor 488 (Fig. 3a), an Alexa Fluor 555 (Fig. 7h), or an Alexa Fluor 633 (Fig. 3b–d and f–h and fig. 7b). Following immunostaining, its image was acquired using either the 488 nm argon-ion laser (Fig. 3a), 543 nm laser (Fig. 7h), or the 633 nm laser (Fig. 3b–d and f–h and Fig. 7b). During image processing, pseudo-colored R-BiP signals (detected in far red) was converted to green (Fig. 3b, g and f) or blue (Fig. 3c, d and h and Fig. 7b). Confocal images were taken using a 510 Meta laser scanning confocal microscope (Zeiss) and analyzed with the Zeiss LSM Image Browser (ver. 4.2.0.121). To determine the intracellular localization of recombinant proteins, cells grown on coverslips were transfected with poly(dA:dT) (1.5 μg/ml). Twenty one hours after transfection, cells were fixed in 4% PFA, followed by the above described procedure.
X-peptide pulldown assay
The X-peptide binding assays were performed essentially as described52. A set of 10-mer BiP-derived peptides (X-EEDKKEDVG-biotin) bearing N-terminal Arg-Glu (permanently arginylated), Glu (native), or Val (Glu-to-Val mutant) residues were cross-linked through C-terminal biotin to streptavidin agarose resin (Thermo Scientific). Alternatively, we used a set of 12-mer peptides (X-I-F-S-T-I-E-G-R-T-Y-K-biotin) in which I-F-S-T-I-E-G-R-T-Y corresponds to residues 2–11 of the Sindbis virus polymerase nsP4 which is degraded according to the N-end rule. The N-terminal X residue was Arg, His, Lys (type-1), Phe, Trp, Tyr, Leu (type-2), Val, Asp or Gly (stabilizing control). The C-terminal biotin moiety was cross-linked to streptavidin agarose resin (Thermo) with a ratio of 0.5 mg peptide per 1 ml settled resin. Following dilution in 5X PBS and incubation with cell extracts overnight at 4°C, the protein-bound beads were collected by centrifugation at 2,000 rpm for 3 min and washed by PBS three times. Protein extracts were prepared using HEK293 cells transiently expressing p62 mutant proteins. Post 24 hrs transfection, cells were collected by centrifugation and lysed in a hypotonic buffer (10 mM HCl, 1.5 mM MgCl2, and 10 mM HEPES, pH 7.9), followed by incubation on ice for 30 min. Cell lysates were obtained through five rounds of freezing-thawing cycles or using dounce homogenization, followed by centrifugation at 13,000 rpm at 4°C for 10 min. For the X-peptide pulldown assay, 150–200 μg total proteins in soluble extracts (30 μl) was diluted in 300 μl binding buffer (0.05% Tween 20, 10% glycerol, 0.2M KCl, and 20mM HEPES, pH 7.9) and mixed with 50 μl (in packed volume) of X-peptide beads. The mixtures were gently rotated at 4°C for 2 hrs. The beads were collected by centrifugation at 2,000 rpm for 30 sec, washed five times each with 1 ml of binding buffer at 4°C for 20 min, resuspended in 20 μl SDS sample buffer, and heated at 100°C for 5 min, followed by SDS-PAGE and immunoblotting.
In vitro p62 oligomerization assays
HEK293 cells were transiently transfected with a plasmid encoding p62-myc/his using Lipofectamine 2000 following manufacturer’s instructions. Approximately 24 hrs after transfection, cells were lysed with cell lysis buffer (50 mM HEPES, pH 7.4, 0.15 M KCl, 0.1% Nonidet P-40, 10% glycerol, and a mixture of protease inhibitors and a phosphatase inhibitor). Following a cycle of freezing and thawing, the cell suspension was incubated on ice for 1 hr and centrifuged at 13,000 x g for 20 min at 4 °C. Protein concentration was determined using the Bradford assay. For p62 oligomerization assays, 1 μg of protein was incubated in the presence or absence of dipeptides and in the presence of 100 μM bestatin at room temperature for 2 hrs. Samples were mixed with a non-reducing loading buffer containing 4% lithium dodecyl sulfate (LDS), heated at 95 °C for 10 min, and resolved on a 3% stacking and 12% separating SDS-PAGE. Immunoblotting analysis using a mixture of anti-p62 and anti-myc antibodies was employed to monitor the conversion of p62 monomers into oligomers and aggregates.
Measurement of p62-LC3 interaction based on the enzyme-linked immunosorbent assay (ELISA)
To determine the specificity of the ELISA, p62−/− MEFs were transfected with a plasmid encoding full length p62 or a truncated p62 fragment (p62-D3) using Lipofectamine 2000. 24 hrs after transfection, cells were harvested and lysed in a lysis buffer (20 mM HEPES, pH7.6, 0.15 M KCl, 0.1% Nonidet P-40, 10% glycerol, and a protease inhibitor cocktail) followed by centrifugation at 13,000 rpm for 20 min at 4°C. Total cell extract (20 μg protein) was incubated with GST-tagged LC3 recombinant protein (Enzo Lifesciences, BML-UW1155) that was immobilized on the glutathione (GSH)-coated plates (Pierce, 15140) in the absence or presence of various concentrations of type-1 and type-2 dipeptides for 1.5 hrs at room temperature. Bound p62 was detected by incubation with anti-p62 antibody for 1 hr at room temperature followed by incubation with HRP-conjugated secondary antibody for 45 min at room temperature. After washing three times with PBS, 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Priece, 34021) was added to each well, and color was developed in the dark at room temperature for 10 min. TMB stop solution, 2 N H2SO4, was added to stop the color reaction. Absorbance was measured on a plate reader at 450 nm.
Statistical analysis
For all experiments shown, n is indicated in the figure legends. Each point value represents the mean ± s.d. from independent experiments unless specified otherwise and depending on the nature of experimental settings. P values were determined using two-way ANOVA (for Fig. 7l) or one-way ANOVA (for Figs. 4f, 6f, and 6g) tests. For the count of R-BiP cytosolic puncta in comparison with those positive for LC3 and p62, at least five different confocal microscopy images were randomly selected, and positive puncta were examined on the images with identical brightness and contrast conditions. The total number of cells on images was determined by using the DAPI (Sigma, D9542) counter staining. Quantification in percentage was performed by counting at least 200 cells per group as indicated in the figure legends. No exclusion criteria were used; data was not excluded. Randomization or blinding of samples was not used in this study. For the representative images (Figs. 1c, 1d, 2a, 2c–q, 3a–h, 4b–e, 5b–c, 5e, 5f, 5h, 5j–l, 6a–c, 6f–h, and 7a–k), the results were reproduced in at least three independent experiments.
Supplementary Material
Acknowledgments
We thank Sung Kyun Ko (KRIBB) for providing HeLa cells stably expressing RFP-GFP-LC3, Suhyun Lee (Seoul National University) for immunoblotting analysis of R-BiP, William T. Kwon (Columbia University) for bioinformatics analysis of N-degrons in the ER, Hyun Jin Jeong (KAIST) for immunostaining analysis of R-BiP and KDEL, Sunna Hong (Yonsei University) for immunostaining analysis of NFκB, and Su Jin Yoo (Middleton High School) for technical assistance. This work was supported by the World Class Institute (WCI) Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (MSIP) of Korea (grant number: WCI 2009-002), KRIBB Research Initiative Program, NIH grant HL083365 (to Y.T.K. and Song Li), the Basic Science Research Programs of the NRF funded by the MSIP (NRF-2013R1A2A2A01014170 to Y.T.K.) and by the Ministry of Education (NRF-2013R1A1A2058983 to Y.D.Y), the Brain Korea 21 PLUS Program (to SNU), the SNU Nobel Laureates Invitation Program (to A.C.), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (AMRF) (to A.C.), and the Israel Science Foundation (ISF) (to A.C.). A.C. is an Israel Cancer Research Fund (ICRF) USA Professor.
Footnotes
AUTHOR CONTRIBUTIONS
Bioinformatic analyses of N-end rule degrons on ER proteins were performed by M.M; antibodies to arginylated ER proteins were generated by D.H.H.; immunoblotting analyses of arginylated ER proteins were performed by K.A.K., Y.D.Y., H.C.-M., K.S.S., J.H., J.K.K. and J.E.Y.; immunostaining of arginylated ER proteins were performed by H.C.-M., J.E.Y., Y.J.L., and N.K.S.; DNA-induce innate immune responses were characterized by K.S.S., H.C.-M., A.Z., S-.H.K., and S.T.K.; the domain of p62 that binds to Nt-Arg was determined by J.M.J. and H.C.-M; the relationship of arginylated ER proteins with misfolded proteins and proteasomal inhibition was investigated by H.C.-M. and S.Y.K.; X-peptide pulldown assay with R-BiP peptides was performed by H.C.-M. and J.E.Y.; and p62 aggregation assay was performed by H.C.-M. and J.E.Y. H.C.-M., K.S.S., J.H, and K.A.K. contributed equally to this work. H.G.L., J.S.A. and B.Y.K. provided guidance, specialized reagents and expertise. Y.T.K., H.C.-M., B.Y.K., and A.C supervised personnel and/or wrote the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Supplementary Information is available in the online version of the paper.
References
- 1.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. Naming a targeting signal. Cell. 1991;64:13–15. doi: 10.1016/0092-8674(91)90202-a. [DOI] [PubMed] [Google Scholar]
- 3.Sriram SM, Kim BY, Kwon YT. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat Rev Mol Cell Biol. 2011;12:735–747. doi: 10.1038/nrm3217. [DOI] [PubMed] [Google Scholar]
- 4.Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–289. doi: 10.1146/annurev-biochem-051710-093308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011;20:1298–1345. doi: 10.1002/pro.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nixon RA. The role of autophagy in neurodegenerative disease. Nature Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 7.Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–528. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Kwon YT, Xia ZX, Davydov IV, Lecker SH, Varshavsky A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3α) of the N-end rule pathway. Mol Cell Biol. 2001;21:8007–8021. doi: 10.1128/MCB.21.23.8007-8021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon YT, et al. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol Cell Biol. 2003;23:8255–8271. doi: 10.1128/MCB.23.22.8255-8271.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tasaki T, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR motif and recognize N degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.An JY, et al. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc Natl Acad Sci USA. 2010;107:1912–1917. doi: 10.1073/pnas.0910267107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tasaki T, et al. UBR box N-recognin-4 (UBR4), an N-recognin of the N-end rule pathway, and its role in yolk sac vascular development and autophagy. Proc Natl Acad Sci USA. 2013;110:3800–3805. doi: 10.1073/pnas.1217358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim ST, et al. The N-end rule proteolytic system in autophagy. Autophagy. 2013;9:1100–1103. doi: 10.4161/auto.24643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tasaki T, et al. The substrate recognition domains of the N-end rule pathway. J Biol Chem. 2009;284:1884–1895. doi: 10.1074/jbc.M803641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwon YT, et al. Mouse and human genes encoding the recognition component of the N-end rule pathway. Proc Natl Acad Sci USA. 1998;95:7898–7903. doi: 10.1073/pnas.95.14.7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sriram SM, Kwon YT. The molecular principles of N-end rule recognition. Nature Struct Mol Biol. 2010;17:1164–1165. doi: 10.1038/nsmb1010-1164. [DOI] [PubMed] [Google Scholar]
- 17.Kwon YT, Kashina AS, Varshavsky A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol Cell Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon YT, et al. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 19.Lee MJ, et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci USA. 2005;102:15030–15035. doi: 10.1073/pnas.0507533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu RG, et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature. 2005;437:981–986. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 21.Lee MJ, et al. Synthetic heterovalent inhibitors targeting recognition E3 components of the N-end rule pathway. Proc Natl Acad Sci USA. 2008;105:100–105. doi: 10.1073/pnas.0708465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee MJ, et al. Characterization of the arginylation branch of the N-end rule pathway in G-protein-mediated proliferation and signaling of cardiomyocytes. J Biol Chem. 2012;287:24043–24052. doi: 10.1074/jbc.M112.364117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim HK, Kim RR, Oh JH, Cho h, Varshavsky A, Hwang CS. The N-terminal methionine of cellular proteins as a degradation signal. Cell. 2014;156:158–169. doi: 10.1016/j.cell.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin-proteasome system. Nature Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 27.Kopito RR. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 28.Ciechanover A. Intracellular protein degradation: from a vague idea through the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Bioorg Med Chem. 2013;21:3400–3410. doi: 10.1016/j.bmc.2013.01.056. [DOI] [PubMed] [Google Scholar]
- 29.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaushik S, Cuervo AM. Chaperone-mediated autophagy: a unique way to enter the lysosome world. Trends Cell Biol. 2012;22:407–417. doi: 10.1016/j.tcb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 32.Wang X, Terpstra EJ. Ubiquitin receptors and protein quality control. J Mol Cell Cardiol. 2013;55:73–84. doi: 10.1016/j.yjmcc.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komatsu M. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 34.Moscat J, Diaz-Meco M. T p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogov V, Dötsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell. 2014;53:167–78. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 36.Filimonenko M, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell. 2010;38:265–279. doi: 10.1016/j.molcel.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 39.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korennykh A, Walter P. Structural basis of the unfolded protein response. Annu Rev Cell Dev Biol. 2012;28:251–277. doi: 10.1146/annurev-cellbio-101011-155826. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Liu R, Ni M, Gill P, Lee AS. Cell surface relocalization of the endoplasmic reticulum chaperone and unfolded protein response regulator GRP78/BiP. J Biol Chem. 2010;285:15065–15075. doi: 10.1074/jbc.M109.087445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coppolino M, et al. Calreticulin is essential for integrin-mediated calcium signalling and cell adhesion. Nature. 1997;386:843–847. doi: 10.1038/386843a0. [DOI] [PubMed] [Google Scholar]
- 43.Scott M, Lu G, Hallett M, Thomas DY. The Hera database and its use in the characterization of endoplasmic reticulum proteins. Bioinformatics. 2004;20:937–944. doi: 10.1093/bioinformatics/bth010. [DOI] [PubMed] [Google Scholar]
- 44.Hu RG, et al. Arginyltransferase, its specificity, putative substrates, bidirectional promoter, and splicing-derived isoforms. J Biol Chem. 2006;281:32559–32573. doi: 10.1074/jbc.M604355200. [DOI] [PubMed] [Google Scholar]
- 45.Decca MB, et al. Post-translational arginylation of calreticulin: a new isospecies of calreticulin component of stress granules. J Biol Chem. 2007;282:8237–8245. doi: 10.1074/jbc.M608559200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitzler TM, Papillon J, Guillemette J, Wing SS, Cybulsky AV. Complement modulates the function of the ubiquitin-proteasome system and endoplasmic reticulum-associated degradation in glomerular epithelial cells. Biochim Biophys Acta. 2012;1823:1007–1016. doi: 10.1016/j.bbamcr.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Pilon M, Schekman R, Romisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plemper RK, Bohmler S, Bordallo J, Sommer T, Wolf DH. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- 49.Hampton RY, Sommer T. Finding the will and the way of ERAD substrate retrotranslocation. Curr Opin Cell Biol. 2012;24:460–466. doi: 10.1016/j.ceb.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 50.Kwon YT, et al. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- 51.Tasaki T, et al. UBR box N-recognin-4 (UBR4), an N-recognin of the N-end rule pathway, and its role in yolk sac vascular development and autophagy. Proc Natl Acad Sci USA. 2013;110:3800–3805. doi: 10.1073/pnas.1217358110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tasaki T, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR motif and recognize N degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.