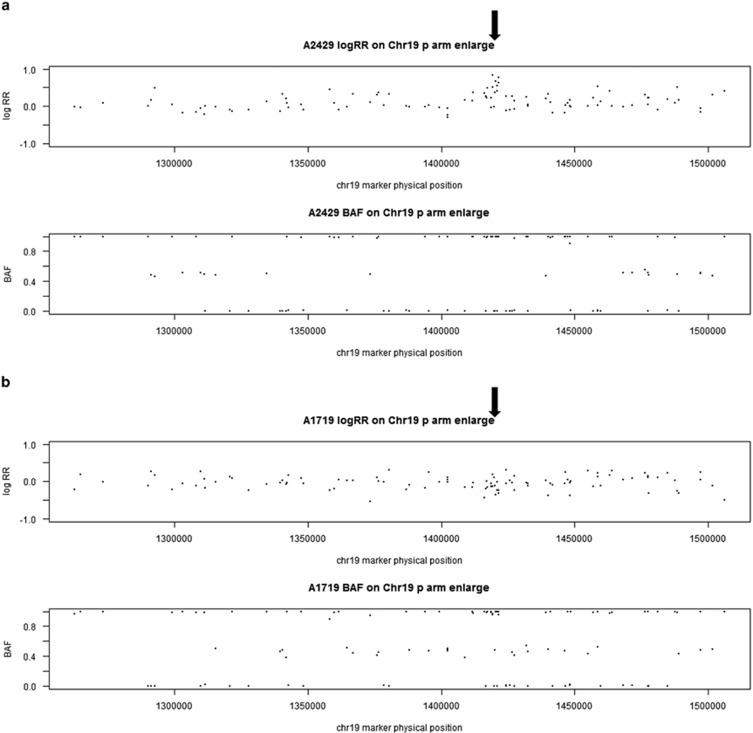
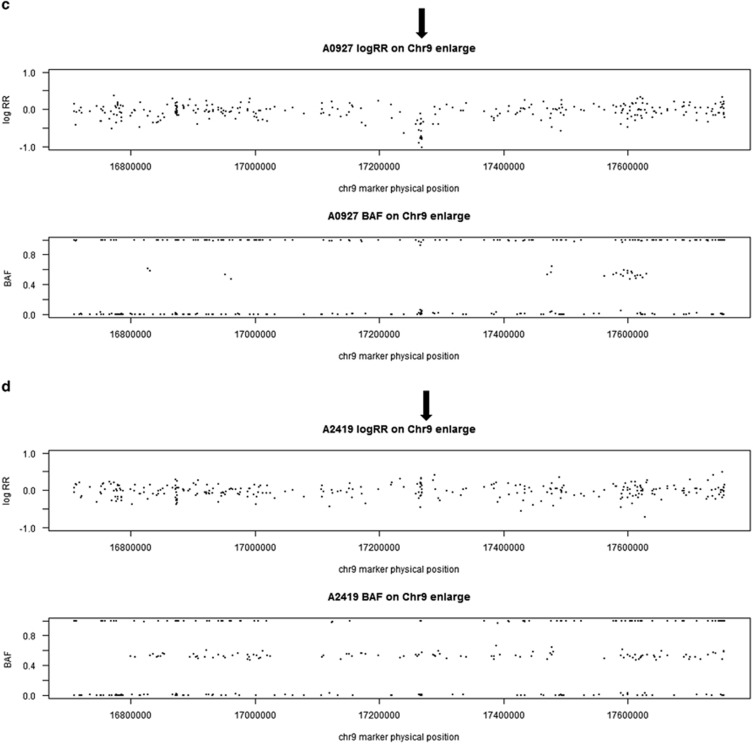
Figure 2.
Examples of duplication on chr 19 in one CNV carrier (a) and one non CNV subject (b), and examples of deletion on chr 9 in one CNV carrier (c) and one non CNV subject (d) based on LRR and BAF. Each data point in the plots is a single marker (SNP or CNV marker). The x axis shows marker's base pair position on chromosome 19, based on Human NCBI Build 36 (hg18). For each subject, the y axis on the top panel is the intensity data termed LRR and the bottom panel is the genotype data termed BAF. (a) Chromosome 19 duplication carrier. In the LRR plot, the expected log2R ratio is zero for normal copy of autosomes; increases in log R ratio were observed in arrowed region; in the BAF plot, the values at 0, 1/2 and 1 represent the expected positions of disomic AA, AB and BB genotypes, respectively. For a hemizygous duplication with copy number of 3, it would have a wider BAF split (at 1/3 and 2/3). However, BAF values at 1/3 and 2/3 were not observed in this case because this region is composed of homozygous SNP markers and a lot of CNV markers. (b) Subject without duplication. In the LRR plot, the arrowed region has LRR around zero; in the BAF plot, the values were clustered around 0, 1/2 and 1 as expected for normal copy number. (c) Chromosome 9 deletion carrier. In the LRR plot, the expected log2R ratio is zero for normal copy of autosomes; decreases in log R ratio were observed in arrowed region; in the BAF plot, the values at 0, 1/2 and 1 represent the expected positions of disomic AA, AB and BB genotypes, respectively. For a hemizygous deletion with copy number of 1, the BAF value on 1/2 disappeared. (d) Subject without chr 9 deletion. In the LRR plot, the arrowed region has LRR around zero; in the BAF plot, the values were clustered around 0, 1/2 and 1 as expected for normal copy number. BAF, B allele frequency; CNV, copy-number variant; LRR, log R ratio; SNP, single-nucleotide polymorphism.